Cdo{rb,py}¶
- Table of contents
- Cdo{rb,py}
Why the ...¶
You're happy with using CDO in a shell? You're hesitant to use a scripting language instead, because shells are so easy to use? Here are some cool features, you'll hardly get within a shell context:
- Direct data access via numpy/narray
If you have python-netcdf4 or scipy installed, you will have direct access to the values of your fields. Python and Ruby offer a rich set of scientific libraries to work with these data arrays, e.g. plotting. - Automatic tempfile handling
When dealing with temporary data, you will never have to do things manually. No cleanup, no manual creation and renaming of files. For whatever you want to save, give a real output file name - the rest is done automatically. - Flexible parallelization
You have a huge long running job, that could be done on parallel? In general shells do not offer enough parallelism to have fine-grained control - the&is not enough for 100 to 1000 jobs, because your system will slow down very much and in a minute, you'll get your call from the administrators.
Both Python and Ruby offer what you just need: Run 1000 routine calls with only 12 concurrent processes or threads. With Cdo{rb,py} you now can make use of it! - Conditional processing
Avoid re-processing if the output files is already on disk. Wisely used, this can speed up your script tremendously. This behaviour can be switched on globally or just for a single calls. E.g. when analysing output data from a running experiments you can run the same script over and over again and you'll always get the latest results. - write new operators out of old ones
Python and Ruby bindings are open libraries, that can be extended at runtime. If you need a new operator, just write it in Python or Ruby!
There is a github repository for easy code sharing and where the changelog is tracked.
If you have questions, please use the CDO forum.
what it is (not) ...¶
This scripting language package is essentially a wrapper around the CDO binary. It parses method arguments and options, builds a command line and executes it. There is no shared library backend which calls CDO operators. This has some advantages:
- operator chaining is fully supported
- multiple CDO binaries can be used at the same time using setCdo())
- packages are highly portable, because they are pure python/ruby implementations
Usage¶
Almost all features are covered by units tests. Theses should be a good starting point to get an impression in how to use the package:
Both bindings are tested with the unix and the win32 version of CDO. Please note, that returning arrays by setting returnCdf is not tested due to the lack of the corresponding netcdf library on windows. There are precompiled windows version of netcdf, but I will not spent time to get it running.
Before doing anything else, the libraries must have been loaded in the usual way:
from cdo import * # python version cdo = Cdo()
In the python version an object has to be created for internal reasons, whereas this is not necessary for Ruby. This may change in the future, but for now it is only a minor difference
require 'cdo' # ruby version
online/offline help¶
For all non-operators, the automatically generated documentation form rubygems might be helpful. Operator documentation can be viewed online, directly by calling
cdo -h <operator pattern>or within the interactive python/ruby shell. Both of the folling examples display the built-in help for sinfov:
- Python:
from cdo import * help(Cdo().sinfov)
orfrom cdo import * cdo = Cdo() help(cdo.sinfov)
- Ruby
require 'cdo' cdo = Cdo.new cdo.help('sinfov') # or Cdo.help(:sinfov)
IO¶
Input and output files can be set with the keywors input and output
cdo.infov(:input => ifile) #ruby version
cdo.showlevel(:input => ifile)
cdo.infov(input=ifile) #python verson
cdo.showlevel(input=ifile)
cdo.timmin(:input => ifile ,:output => ofile) #ruby version
cdo.timmin(input = ifile, output = ofile) #python version
Options¶
Commandline options like '-f' or '-P' can by used via the options keyword:
cdo.timmin(:input => ifile ,:output => ofile,:options => '-f nc') #ruby version
cdo.timmin(input = ifile, output = ofile, options = '-f nc') #python version
Operator arguments have to be given as the first method argument
cdo.remap(gridFile, weightFile,:input => ifile,:output => ofile,:options => '-f nc') #ruby version
cdo.remap(gridFile+","+weighFile, input = ifile, output = ofile, options = '-f nc') #python version
or
cdo.seltimestep('1/10',:input => ifile,:output => ofile,:options => '-r -B F64') #ruby version
cdo.seltimestep('1/10', input = ifile, output = ofile, options = '-r -B F64') #python version
Operator Chains¶
To take real advantage of CDOs internal parallelism, you should work with operator chains as mush as possible:
cdo.setname('random',:input => "-mul -random,r10x10 -enlarge,r10x10 -setyear,2000 -for,1,4",:output => ofile,:options => '-f nc') #ruby version
cdo.setname('random', input = "-mul -random,r10x10 -enlarge,r10x10 -setyear,2000 -for,1,4", output = ofile, options = '-f nc') #python version
Another good example taken from the Tutorial illustrates the different ways of chaining: While the chain
cdo sub -dayavg ifile2 -timavg ifile1 ofileis represented by
cdo.sub(:input => "-dayavg #{ifile2} -timavg #{ifile1}", :output => ofile) #ruby
cdo.sub(input = "-dayavg " + ifile2 + " -timavg " +ifile1, output = ofile) #python
The serial version, which prohibits internal parallelism, creates unnecessary temporal files and is just mentioned for educational reasons would look like
cdo.sub(:input => Cdo.dayavg(:input => ifile2) + " " + Cdo.timavg(:input => ifile1), :output => ofile) #ruby cdo.sub(input = cdo.dayavg(input = ifile2) + " " + cdo.timavg(input = ifile1), output = ofile) #python
or using the
join-method:
cdo.sub(:input => [Cdo.dayavg(:input => ifile2),Cdo.timavg(:input => ifile1)].join(" "), :output => ofile) #ruby
cdo.sub(input = " ".join([cdo.dayavg(input = ifile2),cdo.timavg(input = ifile1)] , output = ofile) #python
Special Features¶
Tempfile handling¶
If the output stream is omitted, a temporary file is written and its name is the return value of the call:
ofile = cdo.timmin(:input => ifile ,:options => '-f nc') #ruby version
ofile = cdo.timmin(input = ifile, options = '-f nc') #python version
Here, the output files are automatically removed, when the scripts finishes. Manual cleanup is not necessary any more unless you encounter a crash. In that case you can use the new (1.4.0) method
cdo.cleanTempDir()or set an alternative directory for storing temporary files with
cdo = Cdo(tempdir='/path/to/new/tempdir') #python cdo = Cdo.new(tempdir: '/path/to/new/tempdir') #ruby
Conditional Processing¶
When processing large number of input files as it is the case in a running experiment, it can be very helpful to suppress the creation of intermediate output if these files are already there. This can speed up your post-processing. In the default behavior, output is created no matter if something is overwritten or not. Conditional processing can be used in two different ways:
- global setting
cdo.forceOutput = False #python
orCdo.forceOutput = false #ruby
This switch changes the default behavior (example)
- operator option
cdo.stdatm("0,10,20",output = ofile, force = False) #pythonorCdo.stdatm(0,10,20,:output => ofile,:force => false) #ruby
The usage of this options allows you to setup the output action very precisely without changing the default (example for good place to uses this feature)
Multi-threadding - Ruby¶
When things can be done in parallel, Python and Ruby offer a smart way to handle this without to much overhead. A Ruby example should illustrate how it can be done: Tutorial
require 'cdo' require 'parallel' iFile = ARGV[0].nil? ? 'ifs_oper_T1279_2011010100.grb' : ARGV[0] targetGridFile = ARGV[1].nil? ? 'cell_grid-r2b07.nc' : ARGV[1] # grid file targetGridweightsFile = ARGV[2].nil? ? 'cell_weight-r2b07.nc' : ARGV[2] # pre-computed interpolation weights nWorkers = ARGV[3].nil? ? 8 : ARGV[3] # number of parallel threads cdo = Cdo.new # lets work in debug mode cdo.debug = true # split the input file wrt to variable names,codes,levels,grids,timesteps,... splitTag = "ifs2icon_skel_split_" cdo.splitname(:input => iFile, :output => splitTag,:options => '-f nc') # collect Files form the split files = Dir.glob("#{splitTag}*.nc") # remap variables in parallel ofiles = Parallel.map(files,:in_processes => nWorkers).each {|file| basename = file[0..-(File.extname(file).size+1)] ofile = cdo.remap(targetGridFile,targetGridweightsFile, :input => file, :output => "remapped_#{basename}.nc") } # Merge all the results together cdo.merge(:input => ofiles.join(" "),:output => 'mergedResults.nc')
In this case the parallelization is done per variable. The only lines, which had to be added for letting the code run on a user defined (see line 7) number of thread are 2, 13, 25, 30 and 32. This approach uses a queue, which takes all jobs and is getting started with q.run. A python version of JobQueue should be easy to implement. Contribution would be appreciated!
Multi-threadding - Python¶
A multiprocessing based example may look like
from cdo import *
import multiprocessing
def showlevel(arg):
return cdo.showlevel(input=arg)
cdo = Cdo()
cdo.debug = True
ifile = '/home/ram/local/data/cdo/GR30L20_fx.nc'
pool = multiprocessing.Pool(1)
results = []
for i in range(0,5):
results.append(pool.apply_async(showlevel, [ifile]))
pool.close()
pool.join()
for res in results:
print(res.get())
Data access via numpy/narray/xarray/cdf¶
When working with netcdf, it is possible to get access to the data in three additional ways:
- a file handle: Using a file handle offers the flexibility to go through the whole file with all it information like variables, dimensions and attributes. To get such an handle form a cdo call, use the
returnCdfkeyword or use thereadCdfmethods:cdo.stdatm("0", options = "-f nc", returnCdf = True).variables["P"][:] #python, access variable 'P' with scipy.io cdo.stdatm(0, :options => "-f nc", :returnCdf => true).var("P").get #ruby , access with ruby-netcdfor return the pure handle withcdo.readCdf(ifile) #python cdo.readCdf(ifile) #ruby
- a numpy/narray object: If a certain variable should be read in, use the
returnArrayinstead ofreturnCdf:pressure = cdo.stdatm("0", options = "-f nc", returnArray = 'P') #python pressure = cdo.stdatm(0, :options => "-f nc", :returnArray = 'P') #ruby - a masked array: If the target variable has missing values, i.e. makes use of the FillValue, the returned structure reflects this. For python a masked array is returned, the ruby version uses a special version of NArray called NArrayMiss. As an example, lets mask out the ocean from the global topography:
oro = cdo.setrtomiss(-10000,0, input = cdo.topo( options = '-f nc'), returnMaArray = 'topo') #python oro = cdo.setrtomiss(-10000,0,:input => cdo.topo(:options => '-f nc'),:returnMaArray => 'topo') #ruby
- netcdf file-handles with
oro = cdo.setrtomiss(-10000,0, input = cdo.topo( options = '-f nc'), returnCdf = True) #python oro = cdo.setrtomiss(-10000,0,:input => cdo.topo(:options => '-f nc'),:returnCdf => true) #ruby
As an python extra there is the option to return XArray/XDataset objects with
oro = cdo.setrtomiss(-10000,0, input = cdo.topo( options = '-f nc'), returnXArray = 'topo') oro = cdo.setrtomiss(-10000,0,:input => cdo.topo(:options => '-f nc'),:returnXDataset = True)
Have a look into the documentation of the underlying netcdf libraries to get an overview of their functionality:
- scipy.io.netcdf (read-only access) or for some examples, look here
- netcdf4-python allows write access, directly from Unidata, recommended
- ruby-netcdf
Prerequisites
The python module requires scipy.io (or pycdf as a fallback) whereas the ruby module needs ruby-netcdf. These dependencies are not handled automatically by pip or gem, because they are optional. Scipy and netcdf4-python are available for most linux/unix distributions as a precompiled package. If this is not the case for your favorite one, you could also use its pip repository. The ruby-netcdf package has a gem-repository:
- Ruby:
gem install ruby-netcdf
orgem install ruby-netcdf --user-install
- Python:
pip install scipy
or visit the the homepage for help on manual installation
Use Cases: Plotting
Examples: Python
from cdo import *
cdo = Cdo() # create the CDO caller
ifile = 'tsurf.nc' # input: surface temperature
cdo.fldsum(input=ifile) # compute the timeseries of global sum, return a temporary filename
vals = cdo.fldsum(input=ifile,returnCdf=True).variables['tsurf'][:] # return the timeseries as numpy array
print(cdo.fldsum(input=ifile,returnCdf=True).variables) # get a list of all variables in the file Basic plotting:
from cdo import *
import matplotlib.pyplot as plt
ifile = 'EH5_AMIP_1_TSURF_1991-1995.nc'
cdo = Cdo()
# Comput the field mean value timeseries and return it as a numpy array
vals = cdo.fldmean(input=ifile,returnCdf=True).variables['tsurf'][:]
# make it 1D
vals = vals.flatten()
# Plot the cumulatice sum of the variataion
plt.plot((vals-vals.mean()).cumsum())
plt.show()
| produces: | original: |
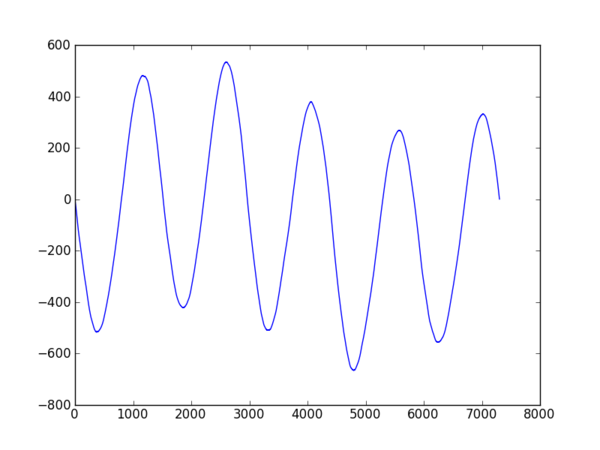 |
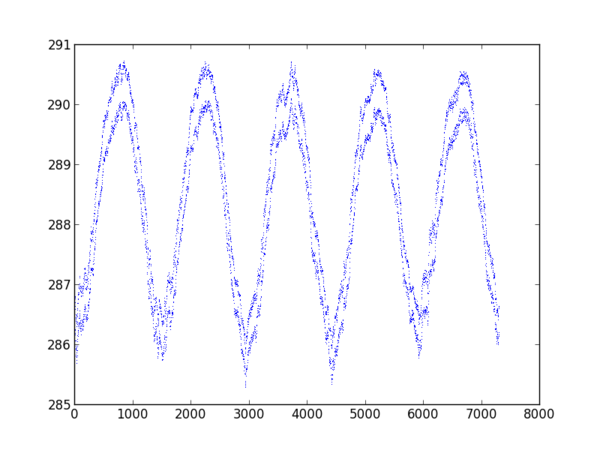 |
2D plotting:
import xarray
from cdo import Cdo
import numpy
from matplotlib import pylab
cdo = Cdo()
# plotting topography with XArray
cdo.topo(returnXArray='topo').plot()
pylab.show()
# ploting based on masked numpy arrays
# orography with missing values below 0
oro = cdo.setrtomiss(-20000,0,input='-sellonlatbox,-20,60,20,60 -topo',returnMaArray='topo')
pylab.imshow(numpy.flipud(oro))
pylab.show()
| XArray | numpy |
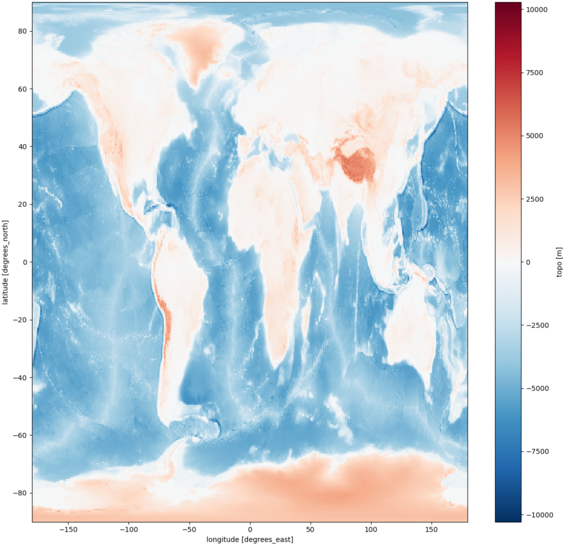 |
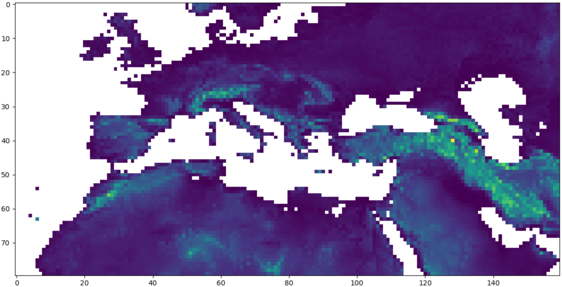 |
Examples: Ruby
require 'cdo'
cdo = Cdo.new
ifile = 'tsurf.nc' # input: surface temperature
vals = cdo.fldsum(:in => ifile,:returnCdf=> true).var('tsurf').get # return the global sum timeseries as narray
puts cdo.fldsum(:input => ifile,:returnCdf => true).var_names # get a list of all variables in the file If you want some basic plotting, use the Ruby bindings of the GNU Scientific Library. You can install it like cdo. Here's a short example:
require 'cdo'
require 'gsl'
cdo = Cdo.new
ifile = "data/examples/EH5_AMIP_1_TSURF_1991-1995.nc"
tmean = cdo.fldmean(:in => ifile,:returnCdf => true).var('tsurf').get
tmean.to_gv.plot("w d title 'AMPI global mean surface temp'")
which shows
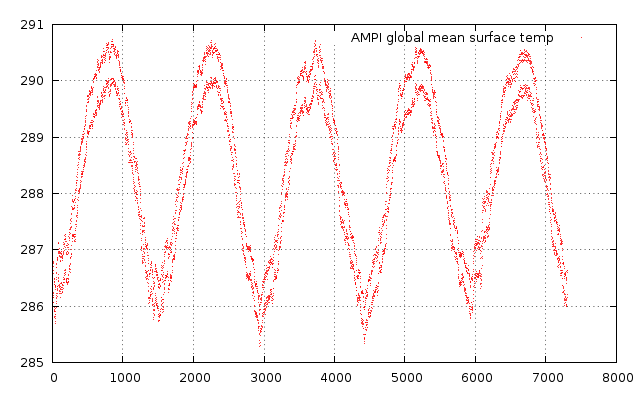
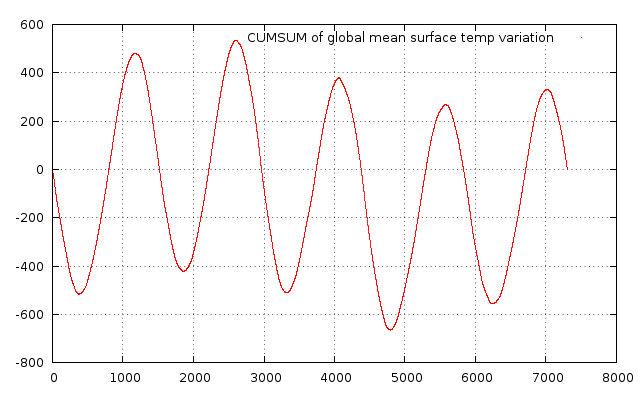
In this context the variable tmean is of type narray which is the ruby version of numpy. It has several methods itself. For filtering the out the temporal behaviour of the aboce time series, you could substract the mean value and display the cumulative sum by adding:
(tmean-tmean.mean)[0,0,0..-1].cumsum.to_gv.plot("w d title 'CUMSUM of global mean surface temp variation'")with results
Use Cases: Interpolation, Root finding, Data fitting, ...
Through the numpy/narray interface, both python and ruby version offer a huge amount of extra functionality via several 3rd party libraries:
- Gnu Scientific Library (python: PyGSL, ruby: rb-gsl)
- scipy (python only) : Large toolset from linear algebra, rbf to 3D vizualization
- R (python , ruby): Statistical computing
Write your own operators¶
Future versions¶
Both cdo modules are not directly linked to a special CDO version. Instead you can change the CDO version to what ever you have installed. Use the setCdo method to use another CDO binary. When CDO is updated and new operators area available, they are usable in the python and ruby modules automatically without any update.
Installation¶
CDO can be easily accessed via Ruby and Python. For each of the these two there is a dedicated package with can be installed from public servers with their own specific package management systems: gem for Ruby and pypi for Python. Interfaces of both packages are the identical.
Note on Windows: There is no native windows-build of CDO, since it is designed for POSIX-compatible systems. Please have a look at Win32 for how to run CDO on windows-based systems. The sort version is: Install cygwin
+ some additional cygwin packages incl. Ruby or Python and use the pre-build cdo package from the download area.
Ruby¶
Ruby's package system is called _gem_. The cdo module is located here. Its installation is rather easy: Just type
gem install cdoand you'll get the latest version installed on your system. Installation as usual requires root privileges, so it might be necessary to prepend a sudo to the command. gem has a great built-in help:
gem help installwill show all you need for installation. If you do not have root access to you machine, another installation directory should be chosen with the
--install-dir option or you use gem install cdo --user-installfor an installation under
$HOME/.gem.
Ruby 1.9.x comes with gem included, but some distros like debian and its derivates create extra packages for it. You might have to watch out for a rubygems package.
Python: pip¶
The cdo module can also be installed for python using pypi, the python package index. Cdo can be found here. If _pip_ is installed on your system, just type
pip install cdoFor user installations, use
pip install cdo --userPlease Note: For upgrading with pip, you have to remove the temporary directories first. Otherwise the upgrade will not take place:
rm -rf /tmp/pip /tmp/src && pip install --build=/tmp/pip --src=/tmp/src --user cdo --upgrade
Without pip, you should download the tar file and run (possibly requiring root privileges)
python setup.py installin the extracted directory.
Python: conda¶
the conda-forge channel has separate packages for the CDO binary and the python bindings:
| CDO binary | python-bindings | |
| conda package name | cdo | python-cdo |
Installation works as usual with
conda -c conda-forge install python-cdo
For JupyterHub users - Watch out for your temporary data¶
In contrast to plain python scripts the JupyterHub environment needs special attentions when using CDO's python bindings.
General handling of intermediate results¶
cdo.py allows to omit output files - instead it will create temporary files for you on the go and clean everything after the script is finished:
>>> ofile = cdo.fldmean(input="myfile.nc")
>>> print(ofile)
/tmp/cdoPyhhp_h4yr
Compared to bash this feature eases the process of writing analysis scripts, because large parts of the boiler plate code is gone.
Normal python script run in order of seconds or minutes. Hence temporary data is removed rather quickly. JupyterHub session on the other hand can run for hours. Further more they run shared with other sessions on the same nodes.
On most Unix-like systems temporary files are stored in main memory. So if there are too many temporary files at the same time the node become unusable had has to be restarted.
How to avoid this?
- Cleanup whenever possible: cdo.py comes with a routine
This will remove all temporary files, which were created by cdo.py and are owned by you. So it will also work on temporary files created by you on that host in a former session.cdo.cleanTempDir()
Add this call in between larger blocks of code, where the temporary data is not needed (e.g. because you used an explicit name for the main results) - Choose your own directory for temporary data right at the start with
With a setup like this you don't interfere with the main memory and have a lot more space. The price you pay is filesystem performance: every normal file system is a lot slower then `/tmp` in main memory. But you get more control and a more stable system.cdo = Cdo(tempdir="/work/mh0287/{}/tmp".format(os.environ['USER']))
Final thoughts¶
JupyterHub is not a normal script because there is no prescribed command execution: cells can be executed top-down, but they don't have to. That's why `cdo.cleanTempDir()` is not a good solution here. If you program using functions, you can add it there, but the interactive nature of the workflow does not ensure the cleanup is run on a regular basis.
My recommendation for JubypterHub is
cdo = Cdo(tempdir="<big-directory>")In my opinion the performance penalty is worth the stability of the system. Technically `cdo.cleanTempDir()` still can be used on whatever tempdir you chose to use, but going away from `/tmp` really helps.