Forums » Operator News »
Masking - more general
Added by Karin Meier-Fleischer over 4 years ago
Create land-sea mask files with CDO¶
- Table of contents
- Create land-sea mask files with CDO
A mask file is represented by missing values (or zeros) and ones in their grid cells. The missing value means that there is no data to be used in the grid cell, and the value one means that the data value within the grid cell has to be used.
CDO offers us a very easy way to create a land-sea mask file of nearly any grid resolution you want by using the operators topo and expr. The operator topo provides us with the integrated topography dataset as a basis to select the ocean or land areas which can then used by the expr operator.
But before we start we should think about what we specifically want as a result, mask land areas or mask ocean areas.
We need:
a) input dataset
b) if necessary, interpolate the input dataset to another grid
c) create a mask file to mask ocean area grid cells for the same grid
d) select the data using the mask file
1. Mask ocean areas (missing values in ocean areas)¶
Let's get started:
a) The input dataset here is called infile.nc
cdo sinfon infile.nc
File format : NetCDF
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Parameter name
1 : MPIMET ECHAM5.2 v instant 17 1 18432 1 F32 : t
Grid coordinates :
1 : gaussian : points=18432 (192x96) F48
lon : -180 to 178.125 by 1.875 degrees_east circular
lat : 88.57217 to -88.57217 degrees_north
Vertical coordinates :
1 : pressure : levels=17
lev : 100000 to 1000 Pa
Time coordinate : 1 step
RefTime = 2001-01-01 00:00:00 Units = hours Calendar = standard
YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss
2001-01-01 00:00:00
b) We want to interpolate the input data to a global 1x1 degree grid with the use of the remapbil operator.
cdo -remapbil,r360x180 infile.nc infile_r360x180.nc
cdo sinfon infile_r360x180.nc
File format : NetCDF
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Parameter name
1 : MPIMET ECHAM5.2 v instant 17 1 64800 1 F32 : t
Grid coordinates :
1 : lonlat : points=64800 (360x180)
lon : 0 to 359 by 1 degrees_east circular
lat : -89.5 to 89.5 by 1 degrees_north
Vertical coordinates :
1 : pressure : levels=17
lev : 100000 to 1000 Pa
Time coordinate : 1 step
RefTime = 2001-01-01 00:00:00 Units = hours Calendar = standard
YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss
2001-01-01 00:00:00
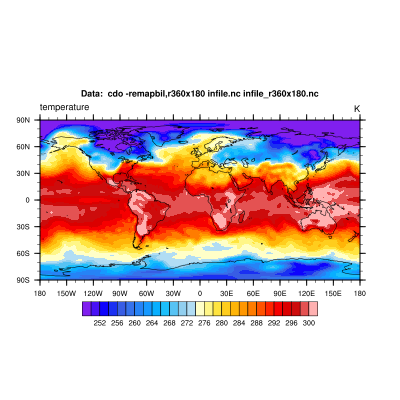
c) Create the mask file by setting the selected grid cells of ocean areas to missing value. We use the topo operator to get the topography data which allows us to select a specific value range of the variable topo. The integrated topography dataset has values in a range of -7791.58 m and 5450.83 m.
Get the topography dataset on a 1x1 degree grid:
cdo -f nc -remapbil,r360x180 -topo topo_r360x180.nc
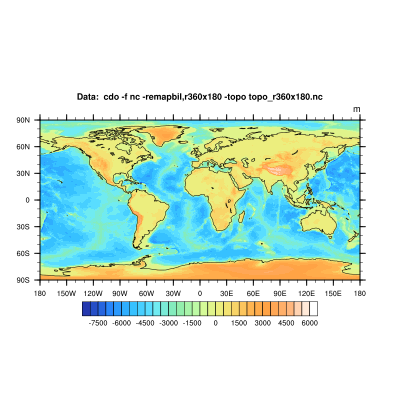
We've interpolated the topography data to the same grid as our interpolated input dataset from above. It is only possible to mask datasets with mask files of the same grid.
Now, we can mask the ocean by setting the topo values between -8000 and 0 m to missing value and the topo data on land areas to 1.
cdo -expr,'topo = ((topo>=0.0)) ? 1.0 : topo/0.0' topo_r360x180.nc mask_ocean.nc
cdo infon mask_ocean.nc
-1 : Date Time Level Gridsize Miss : Minimum Mean Maximum : Parameter name
1 : 0000-00-00 00:00:00 0 64800 43136 : 1.0000 1.0000 1.0000 : topo
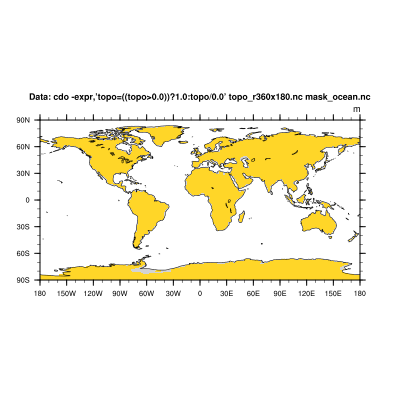
d) In the next step we mask our input dataset with the mask file from above by multiplying them simply.
cdo -mul mask_ocean.nc infile_r360x180.nc infile_r360x180_mask_ocean.nc
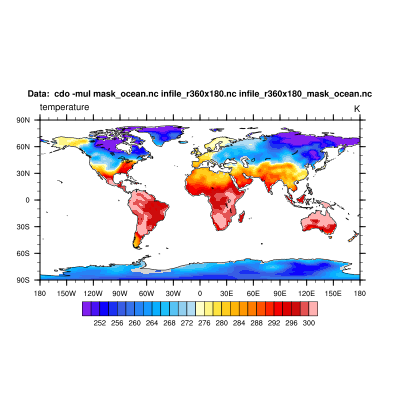
2. Mask land areas (missing values in ocean areas)¶
You need the same as above except for the step to create the mask file. This time we need to set the grid cells for the land areas to missing value, and that's why we have to create another mask file.
c) Create a mask file to be used to mask land area grid cells for the same grid
We can mask the land areas by setting the topo values between 0 and 6000 m to missing value and the topo data on ocean areas to 1.
cdo -expr,'topo = ((topo<0.0)) ? 1.0 : topo/0.0' topo_r360x180.nc mask_land.nc
cdo infon mask_land.nc
-1 : Date Time Level Gridsize Miss : Minimum Mean Maximum : Parameter name
1 : 0000-00-00 00:00:00 0 64800 21667 : 1.0000 1.0000 1.0000 : topo
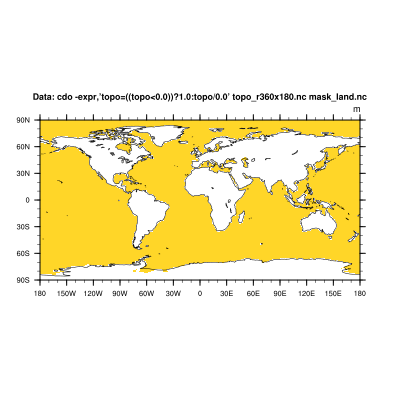
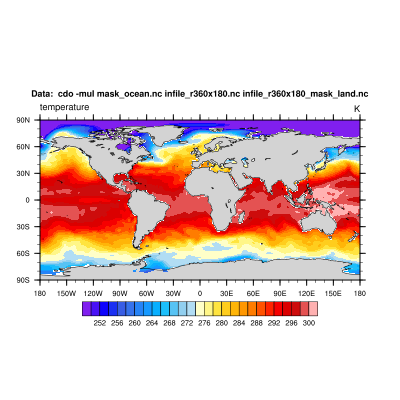
d) And again, we mask our input dataset with the mask file from above by simply multiplying them.
cdo -mul mask_land.nc infile_r360x180.nc infile_r360x180_mask_land.nc
3. Alternative way (set areas to be mask to value 0)¶
Mask ocean areas:¶
c) Create a mask file containing 0 and 1 (0=do not use; 1=use)
cdo -gec,0 topo_r360x180.nc mask_ocean_2.nc
d) Here, we have to divide the input data by the mask file data (the result of division by 0 is missing value).
cdo -div infile_r360x180.nc mask_ocean_2.nc infile_r360x180_mask_ocean_2.nc
Mask land areas:¶
c) Create a mask file containing 0 and 1 (0=not use; 1=use)
cdo -ltc,0 topo_r360x180.nc mask_land_2.nc
d) Again, we have to divide the input data by the mask file data.
cdo -div infile_r360x180.nc mask_land_2.nc infile_r360x180_mask_land_2.nc
4. Make it short¶
The best always comes at the end. 
Now, hopefully it is clear what masking means and that there are different ways to do it. If you are familiar with CDO's operator chaining, then the above described in detail also works in one single command line call.
Mask ocean area in one command line:¶
cdo -f nc -mul -expr,'topo=((topo>0.0))?1.0:topo/0.0' -remapbil,r360x180 -topo infile_r360x180.nc infile_r360x180_mask_ocean.nc
or going the other way
cdo -f nc -div infile_r360x180.nc -gec,0 -remapbil,r360x180 -topo infile_r360x180_mask_ocean.nc
Mask land area in one command line:¶
cdo -f nc -mul -expr,'topo=((topo<0.0))?1.0:topo/0.0' -remapbil,r360x180 -topo infile_r360x180.nc infile_r360x180_mask_ocean.nc
or going the other way
cdo -f nc -div infile_r360x180.nc -ltc,0 -remapbil,r360x180 -topo infile_r360x180_mask_land.nc

| plot_mask.png (39.1 KB) plot_mask.png | |||
| plot_t.png (54.2 KB) plot_t.png | |||
| plot_tmask.png (49.3 KB) plot_tmask.png | |||
| plot_topo.png (64.4 KB) plot_topo.png | |||
| plot_tmask2.png (51.2 KB) plot_tmask2.png | |||
| plot_mask2.png (39.2 KB) plot_mask2.png |
Replies (3)
RE: Masking - more general - Added by Ralf Mueller over 4 years ago
hi Karin, great post!
It perfectly fits to the current situation - everyone is wearing masks 
cheers
ralf
RE: Masking - more general - Added by Pedro Alencar almost 3 years ago
Hi, thanks for the great article!
Only one thing: when I apply the method, in the end the generated nc file has the variable name topo and unit m. The values are what I expected, only the names are changes. is there a way to keep the correct names?
cheers
pedro
RE: Masking - more general - Added by Karin Meier-Fleischer almost 3 years ago
What names are incorrect? Please, be more specific.