CDO User Guide

Climate Data Operator
Version 2.5.4
Nov 2025
Uwe Schulzweida – MPI for Meteorology

Contents
1.1 Installation
1.1.1 Unix
1.1.2 MacOS
1.1.3 Windows
1.2 Usage
1.2.1 Options
1.2.2 Environment variables
1.2.3 Operators
1.2.4 Parallelized operators
1.2.5 Operator parameter
1.2.6 Operator chaining
1.2.7 Chaining Benefits
1.3 Advanced Usage
1.3.1 Wildcards
1.3.2 Argument Groups
1.3.3 Applying a operator or chain to multiple inputs
1.3.4 Apply with [:] notation
1.3.5 Apply Keyword (LEGACY)
1.4 Memory Requirements
1.5 Horizontal grids
1.5.1 Grid area weights
1.5.2 Grid description
1.5.3 ICON - Grid File Server
1.6 Z-axis description
1.7 Time axis
1.7.1 Absolute time
1.7.2 Relative time
1.7.3 Conversion of the time
1.8 Parameter table
1.9 Missing values
1.9.1 Mean and average
1.10 Percentile
1.10.1 Percentile over timesteps
1.11 Regions
2 Reference manual
2.1 Information
2.1.1 INFO - Information and simple statistics
2.1.2 SINFO - Short information
2.1.3 XSINFO - Extra short information
2.1.4 DIFF - Compare two datasets field by field
2.1.5 NINFO - Print the number of parameters, levels or times
2.1.6 SHOWINFO - Show variable information
2.1.7 SHOWATTRIBUTE - Show attributes
2.1.8 FILEDES - Dataset description
2.2 File operations
2.2.1 APPLY - Apply operators
2.2.2 COPY - Copy datasets
2.2.3 TEE - Duplicate a data stream and write it to file
2.2.4 PACK - Pack data
2.2.5 UNPACK - Unpack data
2.2.6 SETCHUNKSPEC - Specify chunking
2.2.7 SETFILTER - Specify filter
2.2.8 BITROUNDING - Bit rounding
2.2.9 REPLACE - Replace variables
2.2.10 DUPLICATE - Duplicates a dataset
2.2.11 MERGEGRID - Merge grid
2.2.12 MERGE - Merge datasets
2.2.13 SPLIT - Split a dataset
2.2.14 SPLITTIME - Split timesteps of a dataset
2.2.15 SPLITSEL - Split selected timesteps
2.2.16 SPLITDATE - Splits a file into dates
2.2.17 DISTGRID - Distribute horizontal grid
2.2.18 COLLGRID - Collect horizontal grid
2.3 Selection
2.3.1 SELECT - Select fields
2.3.2 SELMULTI - Select multiple fields via GRIB1 parameters
2.3.3 SELVAR - Select fields
2.3.4 SELTIME - Select timesteps
2.3.5 SELBOX - Select a box
2.3.6 SELREGION - Select horizontal regions
2.3.7 SELGRIDCELL - Select grid cells
2.3.8 SAMPLEGRID - Resample grid
2.3.9 SELYEARIDX - Select year by index
2.3.10 SELTIMEIDX - Select timestep by index
2.3.11 SELSURFACE - Extract surface
2.4 Conditional selection
2.4.1 COND - Conditional select one field
2.4.2 COND2 - Conditional select two fields
2.4.3 CONDC - Conditional select a constant
2.4.4 MAPREDUCE - Reduce fields to user-defined mask
2.5 Comparison
2.5.1 COMP - Comparison of two fields
2.5.2 COMPC - Comparison of a field with a constant
2.5.3 YMONCOMP - Multi-year monthly comparison
2.5.4 YSEASCOMP - Multi-year seasonal comparison
2.6 Modification
2.6.1 SETATTRIBUTE - Set attributes
2.6.2 SETPARTAB - Set parameter table
2.6.3 SET - Set field info
2.6.4 SETTIME - Set time
2.6.5 CHANGE - Change field header
2.6.6 SETGRID - Set grid information
2.6.7 SETZAXIS - Set z-axis information
2.6.8 INVERT - Invert latitudes
2.6.9 INVERTLEV - Invert levels
2.6.10 SHIFTXY - Shift field
2.6.11 MASKREGION - Mask regions
2.6.12 MASKBOX - Mask a box
2.6.13 SETBOX - Set a box to constant
2.6.14 ENLARGE - Enlarge fields
2.6.15 SETMISS - Set missing value
2.6.16 VERTFILLMISS - Vertical filling of missing values
2.6.17 TIMFILLMISS - Temporal filling of missing values
2.6.18 SETGRIDCELL - Set the value of a grid cell
2.7 Arithmetic
2.7.1 EXPR - Evaluate expressions
2.7.2 MATH - Mathematical functions
2.7.3 ARITHC - Arithmetic with a constant
2.7.4 ARITH - Arithmetic on two datasets
2.7.5 DAYARITH - Daily arithmetic
2.7.6 MONARITH - Monthly arithmetic
2.7.7 YEARARITH - Yearly arithmetic
2.7.8 YHOURARITH - Multi-year hourly arithmetic
2.7.9 YDAYARITH - Multi-year daily arithmetic
2.7.10 YMONARITH - Multi-year monthly arithmetic
2.7.11 YSEASARITH - Multi-year seasonal arithmetic
2.7.12 ARITHDAYS - Arithmetic with days
2.7.13 ARITHLAT - Arithmetic with latitude
2.8 Statistical values
2.8.1 TIMCUMSUM - Cumulative sum over all timesteps
2.8.2 CONSECSTAT - Consecute timestep periods
2.8.3 VARSSTAT - Statistical values over all variables
2.8.4 ENSSTAT - Statistical values over an ensemble
2.8.5 ENSSTAT2 - Statistical values over an ensemble
2.8.6 ENSVAL - Ensemble validation tools
2.8.7 FLDSTAT - Statistical values over a field
2.8.8 ZONSTAT - Zonal statistics
2.8.9 MERSTAT - Meridional statistics
2.8.10 GRIDBOXSTAT - Statistical values over grid boxes
2.8.11 REMAPSTAT - Remaps source points to target cells
2.8.12 VERTSTAT - Vertical statistics
2.8.13 TIMSELSTAT - Time range statistics
2.8.14 TIMSELPCTL - Time range percentile values
2.8.15 RUNSTAT - Running statistics
2.8.16 RUNPCTL - Running percentile values
2.8.17 TIMSTAT - Statistical values over all timesteps
2.8.18 TIMPCTL - Percentile values over all timesteps
2.8.19 HOURSTAT - Hourly statistics
2.8.20 HOURPCTL - Hourly percentile values
2.8.21 DAYSTAT - Daily statistics
2.8.22 DAYPCTL - Daily percentile values
2.8.23 MONSTAT - Monthly statistics
2.8.24 MONPCTL - Monthly percentile values
2.8.25 YEARMONSTAT - Yearly mean from monthly data
2.8.26 YEARSTAT - Yearly statistics
2.8.27 YEARPCTL - Yearly percentile values
2.8.28 SEASSTAT - Seasonal statistics
2.8.29 SEASPCTL - Seasonal percentile values
2.8.30 YHOURSTAT - Multi-year hourly statistics
2.8.31 DHOURSTAT - Multi-day hourly statistics
2.8.32 DMINUTESTAT - Multi-day by the minute statistics
2.8.33 YDAYSTAT - Multi-year daily statistics
2.8.34 YDAYPCTL - Multi-year daily percentile values
2.8.35 YMONSTAT - Multi-year monthly statistics
2.8.36 YMONPCTL - Multi-year monthly percentile values
2.8.37 YSEASSTAT - Multi-year seasonal statistics
2.8.38 YSEASPCTL - Multi-year seasonal percentile values
2.8.39 YDRUNSTAT - Multi-year daily running statistics
2.8.40 YDRUNPCTL - Multi-year daily running percentile values
2.9 Correlation and co.
2.9.1 FLDCOR - Correlation in grid space
2.9.2 TIMCOR - Correlation over time
2.9.3 FLDCOVAR - Covariance in grid space
2.9.4 TIMCOVAR - Covariance over time
2.10 Regression
2.10.1 REGRES - Regression
2.10.2 DETREND - Detrend time series
2.10.3 TREND - Trend of time series
2.10.4 TRENDARITH - Add or subtract a trend
2.11 Eof
2.11.1 EOF - Empirical Orthogonal Functions
2.11.2 EOFCOEFF - Principal coefficients of EOFs
2.12 Interpolation
2.12.1 REMAPBIL - Bilinear interpolation
2.12.2 REMAPBIC - Bicubic interpolation
2.12.3 REMAPNN - Nearest neighbor remapping
2.12.4 REMAPDIS - Distance weighted average remapping
2.12.5 REMAPCON - First order conservative remapping
2.12.6 REMAPLAF - Largest area fraction remapping
2.12.7 REMAP - Grid remapping
2.12.8 REMAPETA - Remap vertical hybrid level
2.12.9 VERTINTML - Vertical interpolation
2.12.10 VERTINTAP - Vertical pressure interpolation
2.12.11 VERTINTGH - Vertical height interpolation
2.12.12 INTLEVEL - Linear level interpolation
2.12.13 INTLEVEL3D - Linear level interpolation from/to 3D vertical coordinates
2.12.14 INTTIME - Time interpolation
2.12.15 INTYEAR - Year interpolation
2.13 Transformation
2.13.1 SPECTRAL - Spectral transformation
2.13.2 SPECCONV - Spectral conversion
2.13.3 WIND2 - D and V to velocity potential and stream function
2.13.4 WIND - Wind transformation
2.13.5 FOURIER - Fourier transformation
2.14 Import/Export
2.14.1 IMPORTBINARY - Import binary data sets
2.14.2 IMPORTCMSAF - Import CM-SAF HDF5 files
2.14.3 INPUT - Formatted input
2.14.4 OUTPUT - Formatted output
2.14.5 OUTPUTTAB - Table output
2.14.6 OUTPUTGMT - GMT output
2.15 Miscellaneous
2.15.1 GRADSDES - GrADS data descriptor file
2.15.2 AFTERBURNER - ECHAM standard post processor
2.15.3 FILTER - Time series filtering
2.15.4 GRIDCELL - Grid cell quantities
2.15.5 SMOOTH - Smooth grid points
2.15.6 DELTAT - Difference between timesteps
2.15.7 REPLACEVALUES - Replace variable values
2.15.8 GETGRIDCELL - Get grid cell index
2.15.9 VARGEN - Generate a field
2.15.10 TIMSORT - Timsort
2.15.11 WINDTRANS - Wind Transformation
2.15.12 ROTUV - Rotation
2.15.13 MROTUVB - Backward rotation of MPIOM data
2.15.14 MASTRFU - Mass stream function
2.15.15 PRESSURE - Pressure on model levels
2.15.16 DERIVEPAR - Derived model parameters
2.15.17 ADISIT - Potential temperature to in-situ temperature and vice versa
2.15.18 RHOPOT - Calculates potential density
2.15.19 HISTOGRAM - Histogram
2.15.20 SETHALO - Set the bounds of a field
2.15.21 WCT - Windchill temperature
2.15.22 FDNS - Frost days where no snow index per time period
2.15.23 STRWIN - Strong wind days index per time period
2.15.24 STRBRE - Strong breeze days index per time period
2.15.25 STRGAL - Strong gale days index per time period
2.15.26 HURR - Hurricane days index per time period
2.15.27 CMORLITE - CMOR lite
2.15.28 VERIFYGRID - Verify grid coordinates
2.15.29 HEALPIX - Change healpix resolution
3 Contributors
3.1 History
3.2 External sources
3.3 Contributors
A Environment Variables
B Parallelized operators
C Standard name table
D Grid description examples
D.1 Example of a curvilinear grid description
D.2 Example description for an unstructured grid
Operator catalog
Operator list
1 Introduction
The Climate Data Operator (CDO) software is a collection of many operators for standard processing of climate and forecast model data. The operators include simple statistical and arithmetic functions, data selection and subsampling tools, and spatial interpolation. CDO was developed to have the same set of processing functions for GRIB [GRIB] and NetCDF [NetCDF] datasets in one package.
The Climate Data Interface [CDI] is used for the fast and file format independent access to GRIB and NetCDF datasets. The local MPI-MET data formats SERVICE, EXTRA and IEG are also supported.
There are some limitations for GRIB and NetCDF datasets:
- GRIB
-
datasets have to be consistent, similar to NetCDF. That means all time steps need to have the same variables, and within a time step each variable may occur only once. Multiple fields in single GRIB2 messages are not supported!
- NetCDF
-
datasets are only supported for the classic data model and arrays up to 4 dimensions. These dimensions should only be used by the horizontal and vertical grid and the time. The NetCDF attributes should follow the GDT, COARDS or CF Conventions.
The main CDO features are:
-
More than 700 operators available
-
Modular design and easily extendable with new operators
-
Very simple UNIX command line interface
-
A dataset can be processed by several operators, without storing the interim results in files
-
Most operators handle datasets with missing values
-
Fast processing of large datasets
-
Support of many different grid types
-
Tested on many UNIX/Linux systems, Cygwin, and MacOS-X
Latest pdf documentation be found here.
1.1 Installation
CDO is supported in different operative systems such as Unix, macOS and Windows. This section describes how to install CDO in those platforms. More examples are found on the main website ( https://code.mpimet.mpg.de/projects/cdo/wiki)
1.1.1 Unix
1.1.1.1. Prebuilt CDO packages
Prebuilt CDO versions are available in online Unix repositories, and you can install them by typing on the Unix terminal
apt-get install cdo
Note that prebuilt libraries do not offer the most recent version, and their version might vary with the Unix
system (see table below). It is recommended to build from the source or Conda environment for an updated
version or a customised setting.
| Unix OS | CDO Version | |
| 11 (Bullseye) | 1.9.10-1 | |
| 10 (Buster) | 1.9.6-1 | |
|
Debian | Sid | 2.0.6-2 |
| 13 | 2.0.6 | |
|
FreeBSD | 12 | 2.0.6 |
| Leap 15.3 | 2.0.6 | |
|
openSUSE | Tumbleweed | 2.0.6-1 |
| 18.04 LTS | 1.9.3 | |
| 20.04 LTS | 1.9.9 | |
|
Ubuntu | 22.04 LTS | 2.0.4-1 |
1.1.1.2. Building from sources
CDO uses the GNU configure and build system for compilation. The only requirement is a working ISO C++20 and C11 compiler.
First go to the download page (https://code.mpimet.mpg.de/projects/cdo) to get the latest distribution, if you do not have it yet.
To take full advantage of CDO features the following additional libraries should be installed:
-
Unidata NetCDF library (https://www.unidata.ucar.edu/software/netcdf) version 4.3.3 or higher.
This library is needed to process NetCDF [NetCDF] files with CDO. -
ECMWF ecCodes library (https://software.ecmwf.int/wiki/display/ECC/ecCodes+Home) version 2.3.0 or higher. This library is needed to process GRIB2 files with CDO.
-
HDF5 szip library (https://www.hdfgroup.org/doc_resource/SZIP) version 2.1 or higher.
This library is needed to process szip compressed GRIB [GRIB] files with CDO. -
HDF5 library (https://www.hdfgroup.org) version 1.6 or higher.
This library is needed to import CM-SAF [CM-SAF] HDF5 files with the CDO operator import_cmsaf. -
PROJ library (https://proj.org) version 5.0 or higher.
This library is needed to convert Sinusoidal and Lambert Azimuthal Equal Area coordinates to geographic coordinates, for e.g. remapping. -
Magics library (https://software.ecmwf.int/wiki/display/MAGP/Magics) version 2.18 or higher.
This library is needed to create contour, vector and graph plots with CDO.
Compilation
Compilation is done by performing the following steps:
-
Unpack the archive, if you haven’t done that yet:
gunzip cdo-$VERSION.tar.gz # uncompress the archive tar xf cdo-$VERSION.tar # unpack it cd cdo-$VERSION -
Run the configure script:
./configure-
Optionaly with NetCDF [NetCDF] support:
./configure --with-netcdf=<NetCDF root directory> -
and with ecCodes:
./configure --with-eccodes=<ecCodes root directory>
For an overview of other configuration options use
./configure --help -
-
Compile the program by running make:
make
The program should compile without problems and the binary (cdo) should be available in the src directory of the distribution.
Installation
After the compilation of the source code do a make install, possibly as root if the destination permissions require that.
make install
The binary is installed into the directory <prefix>/bin. <prefix> defaults to /usr/local but can be changed with the --prefix option of the configure script.
Alternatively, you can also copy the binary from the src directory manually to some bin directory in your search path.
1.1.1.3. Conda
Conda is an open-source package manager and environment management system for various languages (Python, R, etc.). Conda is installed via Anaconda or Miniconda. Unlike Anaconda, miniconda is a lightweight conda distribution. They can be dowloaded from the main conda Website (https://conda.io/projects/conda/en/latest/user-guide/install/linux.html) or on the terminal
wget https://repo.anaconda.com/archive/Anaconda3-2021.11-Linux-x86_64.sh bash Anaconda3-2021.11-Linux-x86_64.sh source ~/.bashrc
and
wget https://repo.continuum.io/miniconda/Miniconda3-latest-Linux-x86_64.sh sh Miniconda3-latest-Linux-x86_64.sh
Upon setting your conda environment, you can install CDO using conda
conda install cdo conda install python-cdo
1.1.2 MacOS
Among the MacOS package managers, CDO can be installed from Homebrew and Macports. The installation via Homebrew is straight forward process on the terminal
brew install cdo
Similarly, Macports
port install cdo
In contrast to Homebrew, Macport allows you to enable GRIB2, szip compression and Magics++ graphic in CDO installation.
port install cdo +grib_api +magicspp +szip
In addition, you could also set CDO via Conda as Unix. You can follow this tutorial to install anaconda or miniconda in your computer (https://conda.io/projects/conda/en/latest/user-guide/install/macos.html). Then, you can install cdo by
conda install -c conda-forge cdo
1.1.3 Windows
Currently, CDO is not supported in Windows system and the binary is not available in the windows conda
repository. Therefore, CDO needs to be set in a virtual environment. Here, it covers the installation of CDO using
Windows Subsystem Linux (WSL) and virtual machines.
1.1.3.1. WSL
WSL emulates Unix in your Windows system. Then, you can install Unix libraries and software such as CDO or the linux conda distribution in your computer. Also, it allows you to directly share your files between your Windows and the WSL environment. However, more complex functions that require a graphic interface are not allowed.
In Windows 10 or newer, WSL can be readily set in your cmd by typing
wsl --install
This command will install, by default, Ubuntu 20.04 in WSL2. You could also choose a different system from this list.
wsl -l -o
Then, you can install your WSL environment as
wsl --install -d NAME
1.1.3.2. Virtual machine
Virtual machines can emulate different operative systems in your computer. Virtual machines are guest
computers mounted inside your host computer. You can set a Linux distribution in your Windows
device in this particular case. The advantages of Virtual machines to WSL are the graphical interface
and the fully operational Linux system. You can follow any tutorial on the internet such as this one
Finally, you can install CDO following any method listed in the section 1.1.1.
1.2 Usage
This section descibes how to use CDO. The syntax is:
cdo [ Options ] Operator1 [ -Operator2 [ -OperatorN ] ]
1.2.1 Options
All options have to be placed before the first operator. The following options are available for all operators:
| -a | Generate an absolute time axis. |
| -b <nbits> | Set the number of bits for the output precision. The valid precisions depend |
| on the file format: |
|
| For srv, ext and ieg format the letter L or B can be added to set the byteorder |
| to Little or Big endian. |
| --cmor | CMOR conform NetCDF output. |
| -C, --color | Colorized output messages. |
| --double | Using double precision floats for data in memory. |
| --eccodes | Use ecCodes to decode/encode GRIB1 messages. |
| --filter <filterspec> |
| NetCDF4 filter specification. |
| -f <format> | Set the output file format. The valid file formats are: |
|
| GRIB2 is only available if CDO was compiled with ecCodes support and all |
| NetCDF file types are only available if CDO was compiled with NetCDF support! |
| -g <grid> | Define the default grid description by name or from file (see chapter 1.3 on page 87). |
| Available grid names are: global_<DXY>, zonal_<DY>, r<NX>x<NY>, lon=<LON>/lat=<LAT>, |
| F<N>, gme<NI>, hpz<ZOOM> |
| -h, --help | Help information for the operators. |
| --no_history | Do not append to NetCDF history global attribute. |
| --netcdf_hdr_pad, --hdr_pad, --header_pad <nbr> |
| Pad NetCDF output header with nbr bytes. |
| -k <chunktype> | NetCDF4 chunk type: auto, grid or lines. |
| -L | Lock I/O (sequential access). |
| -m <missval> | Set the missing value of non NetCDF files (default: -9e+33). |
| -O | Overwrite existing output file, if checked. |
| Existing output file is checked only for: ens<STAT>, merge, mergetime |
| --operators | List of all operators. |
| -P <nthreads> | Set number of OpenMP threads (Only available if OpenMP support was compiled in). |
| --pedantic | Warnings count as errors. |
| --percentile <method> |
| Methods: nrank, nist, rtype8, <NumPy method (linear|lower|higher|nearest|...)> |
| --reduce_dim | Reduce NetCDF dimensions. |
| -R, --regular | Convert GRIB1 data from global reduced to regular Gaussian grid (only with cgribex lib). |
| -r | Generate a relative time axis. |
| -S | Create an extra output stream for the module TIMSTAT. This stream contains |
| the number of non missing values for each output period. |
| -s, --silent | Silent mode. |
| --shuffle | Specify shuffling of variable data bytes before compression (NetCDF). |
| --single | Using single precision floats for data in memory. |
| --sortname | Alphanumeric sorting of NetCDF parameter names. |
| -t <partab> | Set the GRIB1 (cgribex) default parameter table name or file (see chapter 1.6 on page 94). |
| Predefined tables are: echam4 echam5 echam6 mpiom1 ecmwf remo |
| --timestat_date <srcdate> |
| Target timestamp (temporal statistics): first, middle, midhigh or last source timestep. |
| -V, --version | Print the version number. |
| -v, --verbose | Print extra details for some operators. |
| -w | Disable warning messages. |
| --worker <num> | Number of worker to decode/decompress GRIB records. |
| -z aec | AEC compression of GRIB1 records. |
| jpeg | JPEG compression of GRIB2 records. |
| zip[_1-9] | Deflate compression of NetCDF4 variables. |
| zstd[_1-19] | Zstandard compression of NetCDF4 variables. |
1.2.2 Environment variables
There are some environment variables which influence the behavior of CDO. An incomplete list can be found in Appendix A.
Here is an example to set the environment variable CDO_RESET_HISTORY for different shells:
| Bourne shell (sh): | CDO_RESET_HISTORY=1 ; export CDO_RESET_HISTORY |
| Korn shell (ksh): | export CDO_RESET_HISTORY=1 |
| C shell (csh): | setenv CDO_RESET_HISTORY 1 |
1.2.3 Operators
There are more than 700 operators available. A detailed description of all operators can be found in the Reference Manual section.
1.2.4 Parallelized operators
Some of the CDO operators are shared memory parallelized with OpenMP. An OpenMP-enabled C compiler is needed to use this feature. Users may request a specific number of OpenMP threads nthreads with the ’ -P’ switch.
Here is an example to distribute the bilinear interpolation on 8 OpenMP threads:
cdo -P 8 remapbil,targetgrid infile outfile
Many CDO operators are I/O-bound. This means most of the time is spend in reading and writing the data. Only compute intensive CDO operators are parallelized. An incomplete list of OpenMP parallelized operators can be found in Appendix B.
1.2.5 Operator parameter
Some operators need one or more parameter. A list of parameter is indicated by the seperator ’,’.
-
STRING
String parameters require quotes if the string contains blanks or other characters interpreted by the shell. The following command select variables with the name pressure and tsurf:
cdo selvar,pressure,tsurf infile outfile -
FLOAT
Floating point number in any representation. The following command sets the range between 0 and 273.15 of all fields to missing value:
cdo setrtomiss,0,273.15 infile outfile -
BOOL
Boolean parameter in the following representation TRUE/FALSE, T/F or 0/1. To disable the weighting by grid cell area in the calculation of a field mean, use:
cdo fldmean,weights=FALSE infile outfile -
INTEGER
A range of integer parameter can be specified by first/last[/inc]. To select the days 5, 6, 7, 8 and 9 use:
cdo selday,5/9 infile outfileThe result is the same as:
cdo selday,5,6,7,8,9 infile outfile
1.2.6 Operator chaining
Operator chaining allows to combine two or more operators on the command line into a single CDO call. This allows the creation of complex operations out of more simple ones: reductions over several dimensions, file merges and all kinds of analysis processes. All operators with a fixed number of input streams and one output stream can pass the result directly to an other operator. For differentiation between files and operators all operators must be written with a prepended "-" when chaining.
cdo -monmean -add -mulc,2.0 infile1 -daymean infile2 outfile (CDO example call)
Here monmean will have the output of add while add takes the output of mulc,2.0 and daymean. infile1 and infile2 are inputs for their predecessor. When mixing operators with an arbitrary number of input streams extra care needs to be taken. The following examples illustrates why.
-
cdo info -timavg infile1 infile2
-
cdo info -timavg infile?
-
cdo timavg infile1 tmpfile
cdo info tmpfile infile2
rm tmpfile
All three examples produce identical results. The time average will be computed only on the first input
file.
Note(1): In section 1.3.2 we introduce argument groups which will make this a lot easier and less error prone.
Note(2): Operator chaining is implemented over POSIX Threads (pthreads). Therefore this CDO feature is not
available on operating systems without POSIX Threads support!
1.2.7 Chaining Benefits
Combining operators can have several benefits. The most obvious is a performance increase through reducing disk I/O:
cdo sub -dayavg infile2 -timavg infile1 outfile
instead of
cdo timavg infile1 tmp1 cdo dayavg infile2 tmp2 cdo sub tmp2 tmp1 outfile rm tmp1 tmp2
Especially with large input files the reading and writing of intermediate files can have a big influence on the
overall performance.
A second aspect is the execution of operators: Limited by the algorythms potentially all operators of a chain can
run in parallel.
1.3 Advanced Usage
In this section we will introduce advanced features of CDO. These include operator grouping which allows to write more complex CDO calls and the apply keyword which allows to shorten calls that need an operator to be executed on multiple files as well as wildcards which allow to search paths for file signatures. These features have several restrictions and follow rules that depend on the input/output properties. These required properties of operators can be investigated with the following commands which will output a list of operators that have selected properties:
cdo --attribs [arbitrary/filesOnly/onlyFirst/noOutput/obase]
-
arbitrary describes all operators where the number of inputs is not defined.
-
filesOnly are operators that can have other operators as input.
-
onlyFirst shows which operators can only be at the most left position of the polish notation argument chain.
-
noOutput are all operators that do not print to any file (e.g info)
-
obase Here obase describes an operator that does not use the output argument as file but e.g as a file name base (output base). This is almost exclusivly used for operators the split input files.
cdo -splithour baseName_ could result in: baseName_1 baseName_2 ... baseName_N
For checking a single or multiple operator directly the following usage of --attribs can be used:
cdo --attribs operatorName
1.3.1 Wildcards
Wildcards are a standard feature of command line interpreters (shells) on many operating systems. They are
placeholder characters used in file paths that are expanded by the interpreter into file lists. For further information
the Advance Bash Scripting Guide is a valuable source of information. Handling of input is a central issue for
CDO and in some circumstances it is not enough to use the wildcards from the shell. That’s why CDO can handle
them on its own.
| all files | 2020-2-01.txt 2020-2-11.txt 2020-2-15.txt 2020-3-01.txt 2020-3-02.txt |
| 2020-3-12.txt 2020-3-13.txt 2020-3-15.txt 2021.grb 2022.grb | |
| wildcard | filelist results |
| 2020-3* and 2020-3-??.txt | 2020-3-01.txt 2020-3-02.txt 2020-3-12.txt 2020-3-13.txt 2020-3-15.txt |
| 2020-3-?1.txt | 2020-3-01.txt |
| *.grb | 2021.grb 2020.grb |
Use single quotes if the input stream names matched to a single wildcard expression. In this case CDO will do the pattern matching and the output can be combined with other operators. Here is an example for this feature:
cdo timavg -select,name=temperature ’infile?’ outfile
In earlier versions of CDO this was necessary to have the right files parsed to the right operator. Newer
version support this with the argument grouping feature (see 1.3.2). We advice the use of the grouping
mechanism instead of the single quoted wildcards since this feature could be deprecated in future versions.
Note: Wildcard expansion is not available on operating systems without the glob() function!
1.3.2 Argument Groups
In section 1.2.6 we described that it is not possible to chain operators with an arbitrary number of inputs. In this
section we want to show how this can be achieved through the use of operator grouping with angled brackets [].
Using these brackets CDO can assigned the inputs to their corresponding operators during the execution of the
command line. The ability to write operator combination in a parenthis-free way is partly given up in favor of
allowing operators with arbitrary number of inputs. This allows a much more compact way to handle large number
of input files.
The following example shows an example which we will transform from a non-working solution to a working
one.
cdo -infon -div -fldmean -cat infileA -mulc,2.0 infileB -fldmax infileC
This example will throw the following error:
cdo (Abort): -infon -div -fldmean -cat infileA -mulc,2.0 infileB -fldmax infileC ^ Operator cannot be assigned. Reason: Multiple variable input operators used. Use subgroups via [ ] to clarify relations (help: --argument_groups).
The error is raised by the operator div. This operator needs two input streams and one output stream, but the cat operator has claimed all possible streams on its right hand side as input because it accepts an arbitrary number of inputs. Hence it didn’t leave anything for the remaining input or output streams of div. For this we can declare a group which will be passed to the operator left of the group.
cdo -infon -div -fldmean -cat [ infileA -mulc,2.0 infileB ] -fldmax infileC
For full flexibility it is possible to have groups inside groups:
cdo -infon -div -fldmean -cat [ infileA infileB -merge [ infileC1 infileC2 ] ] -fldmax infileD
1.3.3 Applying a operator or chain to multiple inputs
When working with medium or large number of similar files there is a common problem of a processing step (often a reduction) which needs to be performed on all of them before a more specific analysis can be applied. Usually this can be done in two ways: One option is to use a merge operator to glue everything together and chain the reduction step after it. The second option is to write a for-loop over all inputs which perform the basic processing on each of the files separately and call a merge operator one the results. Unfortunately both options have side-effects: The first one needs a lot of memory because all files are read in completely and reduced afterwards while the latter one creates a lot of temporary files. Both memory and disk IO can be bottlenecks and should be avoided. In CDO there exist two approaches to circumvent most drawbacks. The first is to use the more recent ’apply’ feature using the [ to_be_applied : applied_to ] syntax, the second is an older approach and is only listed and documented for completeness sake. We highly recommend the more recent approach!
1.3.4 Apply with [:] notation
With the [ to_be_applied : applied_to ] syntax it is possible to prepend multiple inputs or chains with another chain. For example:
-mergetime [ -selname,tsurf : *.grb ]
would merge all grib files in the folder after selecting the variable tsurf from them.
In general the to_be_applied is applied in parallel to all related input streams (applied_to) before all streams are
passed to operator next in the chain.
The following is an example with three input files:
cdo -mergetime [ -selname,tsurf : infile1 infile2 infile3 ] outfile
This would result in CDO executing as if the following was used:
cdo -mergetime -selname,tsurf infile1 -selname,tsurf infile2 -selname,tsurf infile3 outfile
| Figure 1.1.: | Usage and result of [:] notation |
This notation is especially useful when combined with wildcards. The previous example can be shortened further.
cdo -mergetime [ -selname,tsurf : infile? ] outfile
As shown this feature allows to simplify commands with medium amount of files and to move reductions further back. This can also have a positive impact on the performance.
An example where performance can take a hit.
cdo -yearmean -selname,tsurf -mergetime [ f1 ... f40 ]
An improved but ugly to write example.
cdo -yearmean -mergetime [ -selname,tsurf f1 -selname,tsurf f2 ... -selname,tsurf f40 ]
Apply saves the day. And creates the call above with much less typing.
cdo -yearmean -mergetime [ -selname,tsurf : f1 ... f40 ]
| Figure 1.2.: | [ : ] notation simplifies command and execution |
Further this notation allows to prepend full cdo chains with other operators:
cdo -info -mergetime [ -selname,tsurf : -addc,1 -mul f1 f2 -addc,4 f3 ]
Resolving internally to the following command:
info -mergetime -selname,tsurf [ -addc,1 -mul f1 f2 -selvar,tsurf -addc,4 f3 ]
1.3.5 Apply Keyword (LEGACY)
Originally the apply keyword was introduced for that purpose. We want to repeat that this is an older method and
should not be used in newly written CDO commands! It can be used as an operator, but it needs at least one
operator as a parameter, which is applied in parallel to all related input streams in a parallel way before all
streams are passed to operator next in the chain.
The following is an example with three input files:
cdo -mergetime -apply,-selname,tsurf [ infile1 infile2 infile3 ] outfile
would result in:
cdo -mergetime -selname,tsurf infile1 -selname,tsurf infile2 -selname,tsurf infile3 outfile
| Figure 1.3.: | Usage and result of apply keyword |
Apply is especially useful when combined with wildcards. The previous example can be shortened further.
cdo -mergetime -apply,-selname,tsurf [ infile? ] outfile
As shown this feature allows to simplify commands with medium amount of files and to move reductions further back. This can also have a positive impact on the performance.
An example where performance can take a hit.
cdo -yearmean -selname,tsurf -mergetime [ f1 ... f40 ]
An improved but ugly to write example.
cdo -yearmean -mergetime [ -selname,tsurf f1 -selname,tsurf f2 ... -selname,tsurf f40 ]
Apply saves the day. And creates the call above with much less typing.
cdo -yearmean -mergetime [ -apply,-selname,tsurf [ f1 ... f40 ] ]
| Figure 1.4.: | Apply keyword simplifies command and execution |
In the example in figure 1.4 the resulting call will dramatically save process interaction as well as execution times since the reduction (selname,tsurf) is applied on the files first. That means that the mergetime operator will receive the reduced files and the operations for merging the whole data is saved. For other CDO calls further improvements can be made by adding more arguments to apply (1.5)
A less performant example.
cdo -aReduction -anotherReduction -selname,tsurf -mergetime [ f1 ... f40 ]
cdo -mergetime -apply,"-aReduction -anotherReduction -selname,tsurf" [ f1 ... f40 ]
| Figure 1.5.: | Multi argument apply |
Restrictions: While the apply keyword can be extremely helpful it has several restrictions (for now!).
-
Apply inputs can only be files, wildcards and operators that have 0 inputs and 1 output.
-
Apply can not be used as the first CDO operator.
-
Apply arguments can only be operators with 1 input and 1 output.
-
Grouping inside the Apply argument or input is not allowed.
1.4 Memory Requirements
This section roughly describes the memory requirements of CDO. CDO tries to use as little memory as possible. The smallest unit that is read by all operators is a horizontal field. The required memory depends mainly on the used operators, the data format, the data type and the size of the fields.
The operators have partly very different memory requirements. Many CDO modules like FLDSTAT process one horizontal field at a time. Memory-intensive modules such as ENSSTAT and TIMSTAT require all fields of a time step to be held in memory. Of course, the memory requirements of each operator add up when they are combined. Some operators are parallelized with OpenMP. In multi-threaded mode (see option -P) the memory requirement can increase for these operators. This increase grows with the number of threads used.
The data type determines the number of bytes per value. Single precision floating point data occupies 4 bytes per value. All other data types are read as double precision floats and thus occupy 8 bytes per value. With the CDO option --single all data is read as single precision floats. This can reduce the memory requirement by a factor of 2.
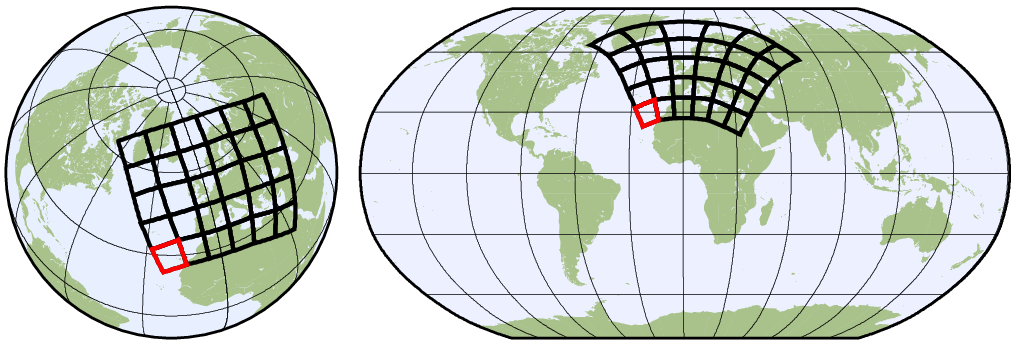
1.5 Horizontal grids
Physical quantities of climate models are typically stored on a horizonal grid. CDO supports structured grids like regular lon/lat or curvilinear grids and also unstructured grids.
1.5.1 Grid area weights
One single point of a horizontal grid represents the mean of a grid cell. These grid cells are typically of different sizes, because the grid points are of varying distance.
Area weights are individual weights for each grid cell. They are needed to compute the area weighted mean or variance of a set of grid cells (e.g. fldmean - the mean value of all grid cells). In CDO the area weights are derived from the grid cell area. If the cell area is not available then it will be computed from the geographical coordinates via spherical triangles. This is only possible if the geographical coordinates of the grid cell corners are available or derivable. Otherwise CDO gives a warning message and uses constant area weights for all grid cells.
The cell area is read automatically from a NetCDF input file if a variable has the corresponding “cell_measures” attribute, e.g.:
var:cell_measures = "area: cell_area" ;
If the computed cell area is not desired then the CDO operator setgridarea can be used to set or overwrite the grid cell area.
1.5.2 Grid description
In the following situations it is necessary to give a description of a horizontal grid:
-
Changing the grid description (operator: setgrid)
-
Horizontal interpolation (all remapping operators)
-
Generating of variables (operator: const, random)
As now described, there are several possibilities to define a horizontal grid.
1.5.2.1. Predefined grids
Predefined grids are available for global regular, gaussian, HEALPix or icosahedral-hexagonal GME grids.
Global regular grid: global_<DXY>
global_<DXY> defines a global regular lon/lat grid. The grid increment <DXY> can be chosen arbitrarily. The longitudes start at <DXY>/2 - 180∘ and the latitudes start at <DXY>/2 - 90∘.
Regional regular grid: dcw:<CountryCode>[_<DXY>]
dcw:<CountryCode>[_<DXY>] defines a regional regular lon/lat grid from the country code. The default value of the optional grid increment <DXY> is 0.1 degree. The ISO two-letter country codes can be found on https://en.wikipedia.org/wiki/ISO_3166-1_alpha-2. To define a state, append the state code to the country code, e.g. USAK for Alaska. For the coordinates of a country CDO uses the DCW (Digital Chart of the World) dataset from GMT. This dataset must be installed on the system and the environment variable DIR_DCW must point to it.
Zonal latitudes: zonal_<DY>
zonal_<DY> defines a grid with zonal latitudes only. The latitude increment <DY> can be chosen arbitrarily. The latitudes start at <DY>/2 - 90∘. The boundaries of each latitude are also generated. The number of longitudes is 1. A grid description of this type is needed to calculate the zonal mean (zonmean) for data on an unstructured grid.
Global regular grid: r<NX>x<NY>
r<NX>x<NY> defines a global regular lon/lat grid. The number of the longitudes <NX> and the latitudes <NY> can be chosen arbitrarily. The longitudes start at 0∘ with an increment of (360/<NX>)∘. The latitudes go from south to north with an increment of (180/<NY>)∘.
One grid point: lon=<LON>/lat=<LAT>
lon=<LON>/lat=<LAT> defines a lon/lat grid with only one grid point.
Full regular Gaussian grid: F<N>
F<N> defines a global regular Gaussian grid. N specifies the number of latitudes lines between the Pole and the Equator. The total number of latitudes and longitues is: nlat = N * 2; nlon = nlat * 2. The longitudes start at 0∘ with an increment of (360/nlon)∘. The gaussian latitudes go from north to south.
Global icosahedral-hexagonal GME grid: gme<NI>
gme<NI> defines a global icosahedral-hexagonal GME grid. NI specifies the number of intervals on a main triangle side.
HEALPix grid: hp<NSIDE>[_<ORDER>]
HEALPix is an acronym for Hierarchical Equal Area isoLatitude Pixelization of a sphere.
hp<NSIDE>[_<ORDER>] defines the parameter of a global HEALPix grid. The NSIDE parameter controls the
resolution of the pixellization. It is the number of pixels on the side of each of the 12 top-level HEALPix pixels.
The total number of grid pixels is 12*NSIDE*NSIDE. NSIDE=1 generates the 12 (H=4, K=3) equal sized top-level
HEALPix pixels. ORDER sets the index ordering convention of the pixels, available are nested (default) or ring
ordering. A shortcut for hp<NSIDE>_nested is hpz<ZOOM>. ZOOM is the zoom level and the relation to NSIDE is
zoom = log2(nside).
If the geographical coordinates are required in CDO, they are calculated from the HEALPix parameters. For this calculation the astropy-healpix C library is used.
1.5.2.2. Grids from data files
You can use the grid description from an other datafile. The format of the datafile and the grid of the data field must be supported by CDO. Use the operator ’sinfo’ to get short informations about your variables and the grids. If there are more then one grid in the datafile the grid description of the first variable will be used. Add the extension :N to the name of the datafile to select grid number N.
1.5.2.3. SCRIP grids
SCRIP (Spherical Coordinate Remapping and Interpolation Package) uses a common grid description for curvilinear and unstructured grids. For more information about the convention see [SCRIP]. This grid description is stored in NetCDF. Therefor it is only available if CDO was compiled with NetCDF support!
SCRIP grid description example of a curvilinear MPIOM [MPIOM] GROB3 grid (only the NetCDF header):
netcdf grob3s { dimensions: grid_size = 12120 ; grid_corners = 4 ; grid_rank = 2 ; variables: int grid_dims(grid_rank) ; double grid_center_lat(grid_size) ; grid_center_lat:units = "degrees" ; grid_center_lat:bounds = "grid_corner_lat" ; double grid_center_lon(grid_size) ; grid_center_lon:units = "degrees" ; grid_center_lon:bounds = "grid_corner_lon" ; int grid_imask(grid_size) ; grid_imask:units = "unitless" ; grid_imask:coordinates = "grid_center_lon grid_center_lat" ; double grid_corner_lat(grid_size, grid_corners) ; grid_corner_lat:units = "degrees" ; double grid_corner_lon(grid_size, grid_corners) ; grid_corner_lon:units = "degrees" ; // global attributes: :title = "grob3s" ; }
1.5.2.4. CDO grids
All supported grids can also be described with the CDO grid description. The following keywords can be used to describe a grid:
| Keyword | Datatype | Description |
| gridtype | STRING | Type of the grid (gaussian, lonlat, curvilinear, unstructured). |
| gridsize | INTEGER | Size of the grid. |
| xsize | INTEGER | Size in x direction (number of longitudes). |
| ysize | INTEGER | Size in y direction (number of latitudes). |
| xvals | FLOAT ARRAY | X values of the grid cell center. |
| yvals | FLOAT ARRAY | Y values of the grid cell center. |
| nvertex | INTEGER | Number of the vertices for all grid cells. |
| xbounds | FLOAT ARRAY | X bounds of each gridbox. |
| ybounds | FLOAT ARRAY | Y bounds of each gridbox. |
| xfirst, xinc | FLOAT, FLOAT | Macros to define xvals with a constant increment, |
| xfirst is the x value of the first grid cell center. | ||
| yfirst, yinc | FLOAT, FLOAT | Macros to define yvals with a constant increment, |
| yfirst is the y value of the first grid cell center. | ||
| xunits | STRING | units of the x axis |
| yunits | STRING | units of the y axis |
Which keywords are necessary depends on the gridtype. The following table gives an overview of the default values or the size with respect to the different grid types.
| gridtype | lonlat | gaussian | projection | curvilinear | unstructured |
| gridsize | xsize*ysize | xsize*ysize | xsize*ysize | xsize*ysize | ncell |
| xsize | nlon | nlon | nx | nlon | gridsize |
| ysize | nlat | nlat | ny | nlat | gridsize |
| xvals | xsize | xsize | xsize | gridsize | gridsize |
| yvals | ysize | ysize | ysize | gridsize | gridsize |
| nvertex | 2 | 2 | 2 | 4 | nv |
| xbounds | 2*xsize | 2*xsize | 2*xsize | 4*gridsize | nv*gridsize |
| ybounds | 2*ysize | 2*ysize | 2*xsize | 4*gridsize | nv*gridsize |
| xunits | degrees | degrees | m | degrees | degrees |
| yunits | degrees | degrees | m | degrees | degrees |
The keywords nvertex, xbounds and ybounds are optional if area weights are not needed. The grid cell corners xbounds and ybounds have to rotate counterclockwise.
CDO grid description example of a T21 gaussian grid:
gridtype = gaussian xsize = 64 ysize = 32 xfirst = 0 xinc = 5.625 yvals = 85.76 80.27 74.75 69.21 63.68 58.14 52.61 47.07 41.53 36.00 30.46 24.92 19.38 13.84 8.31 2.77 -2.77 -8.31 -13.84 -19.38 -24.92 -30.46 -36.00 -41.53 -47.07 -52.61 -58.14 -63.68 -69.21 -74.75 -80.27 -85.76
CDO grid description example of a global regular grid with 60x30 points:
gridtype = lonlat xsize = 60 ysize = 30 xfirst = -177 xinc = 6 yfirst = -87 yinc = 6
The description for a projection is somewhat more complicated. Use the first section to describe the coordinates of the projection with the above keywords. Add the keyword grid_mapping_name to descibe the mapping between the given coordinates and the true latitude and longitude coordinates. grid_mapping_name takes a string value that contains the name of the projection. A list of attributes can be added to define the mapping. The name of the attributes depend on the projection. The valid names of the projection and there attributes follow the NetCDF CF-Convention.
CDO supports the special grid mapping attribute proj_params. These parameter will be passed directly to the PROJ library to generate the geographic coordinates if needed.
The geographic coordinates of the following projections can be generated without the attribute proj_params, if all other attributes are available:
-
rotated_latitude_longitude
-
lambert_conformal_conic
-
lambert_azimuthal_equal_area
-
sinusoidal
-
polar_stereographic
It is recommend to set the attribute proj_params also for the above projections to make sure all PROJ parameter are set correctly.
Here is an example of a CDO grid description using the attribute proj_params to define the PROJ parameter of a polar stereographic projection:
gridtype = projection xsize = 11 ysize = 11 xunits = "meter" yunits = "meter" xfirst = -638000 xinc = 150 yfirst = -3349350 yinc = 150 grid_mapping = crs grid_mapping_name = polar_stereographic proj_params = "+proj=stere +lon_0=-45 +lat_ts=70 +lat_0=90 +x_0=0 +y_0=0"
The result is the same as using the CF conform Grid Mapping Attributes:
gridtype = projection xsize = 11 ysize = 11 xunits = "meter" yunits = "meter" xfirst = -638000 xinc = 150 yfirst = -3349350 yinc = 150 grid_mapping = crs grid_mapping_name = polar_stereographic straight_vertical_longitude_from_pole = -45. standard_parallel = 70. latitude_of_projection_origin = 90. false_easting = 0. false_northing = 0.
CDO grid description example of a regional rotated lon/lat grid:
gridtype = projection xsize = 81 ysize = 91 xunits = "degrees" yunits = "degrees" xfirst = -19.5 xinc = 0.5 yfirst = -25.0 yinc = 0.5 grid_mapping_name = rotated_latitude_longitude grid_north_pole_longitude = -170 grid_north_pole_latitude = 32.5
Example CDO descriptions of a curvilinear and an unstructured grid can be found in Appendix D.
1.5.3 ICON - Grid File Server
The geographic coordinates of the ICON model are located on an unstructured grid. This grid is stored in a separate grid file independent of the model data. The grid files are made available to the general public via a file server. Furthermore, these grid files are located at DKRZ under /pool/data/ICON/grids.
With the CDO function setgrid,<gridfile> this grid information can be added to the data if needed. Here is an example:
cdo sellonlatbox,-20,60,10,70 -setgrid,<path_to_gridfile> icondatafile result
ICON model data in NetCDF format contains the global attribute grid_file_uri. This attribute contains a link to the appropriate grid file on the ICON grid file server. If the global attribute grid_file_uri is present and valid, the grid information can be added automatically. The setgrid function is then no longer required. The environment variable CDO_DOWNLOAD_PATH can be used to select a directory for storing the grid file. If this environment variable is set, the grid file will be automatically downloaded from the grid file server to this directory if needed. If the grid file already exists in the current directory, the environment variable does not need to be set.
If the grid files are available locally, like at DKRZ, they do not need to be fetched from the grid file server. Use the environment variable CDO_ICON_GRIDS to set the root directory of the ICON grids. Here is an example for the ICON grids at DKRZ:
CDO_ICON_GRIDS=/pool/data/ICON
1.6 Z-axis description
Sometimes it is necessary to change the description of a z-axis. This can be done with the operator setzaxis. This operator needs an ASCII formatted file with the description of the z-axis. The following keywords can be used to describe a z-axis:
| Keyword | Datatype | Description |
| zaxistype | STRING | type of the z-axis |
| size | INTEGER | number of levels |
| levels | FLOAT ARRAY | values of the levels |
| lbounds | FLOAT ARRAY | lower level bounds |
| ubounds | FLOAT ARRAY | upper level bounds |
| vctsize | INTEGER | number of vertical coordinate parameters |
| vct | FLOAT ARRAY | vertical coordinate table |
The keywords lbounds and ubounds are optional. vctsize and vct are only necessary to define hybrid model levels.
Available z-axis types:
| Z-axis type | Description | Units |
| surface | Surface | |
| pressure | Pressure level | pascal |
| hybrid | Hybrid model level | |
| height | Height above ground | meter |
| depth_below_sea | Depth below sea level | meter |
| depth_below_land | Depth below land surface | centimeter |
| isentropic | Isentropic (theta) level | kelvin |
Z-axis description example for pressure levels 100, 200, 500, 850 and 1000 hPa:
zaxistype = pressure size = 5 levels = 10000 20000 50000 85000 100000
Z-axis description example for ECHAM5 L19 hybrid model levels:
zaxistype = hybrid size = 19 levels = 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 vctsize = 40 vct = 0 2000 4000 6046.10938 8267.92578 10609.5117 12851.1016 14698.5 15861.125 16116.2383 15356.9258 13621.4609 11101.5625 8127.14453 5125.14062 2549.96875 783.195068 0 0 0 0 0 0 0.000338993268 0.00335718691 0.0130700432 0.0340771675 0.0706498027 0.12591666 0.201195419 0.295519829 0.405408859 0.524931908 0.646107674 0.759697914 0.856437683 0.928747177 0.972985268 0.992281914 1
Note that the vctsize is twice the number of levels plus two and the vertical coordinate table must be specified for the level interfaces.
1.7 Time axis
A time axis describes the time for every timestep. Two time axis types are available: absolute time and relative time axis. CDO tries to maintain the actual type of the time axis for all operators.
1.7.1 Absolute time
An absolute time axis has the current time to each time step. It can be used without knowledge of the calendar. This is preferably used by climate models. In NetCDF files the absolute time axis is represented by the unit of the time: "day as %Y%m%d.%f".
1.7.2 Relative time
A relative time is the time relative to a fixed reference time. The current time results from the reference time and the elapsed interval. The result depends on the calendar used. CDO supports the standard Gregorian, proleptic Gregorian, 360 days, 365 days and 366 days calendars. The relative time axis is preferably used by numerical weather prediction models. In NetCDF files the relative time axis is represented by the unit of the time: "time-units since reference-time", e.g "days since 1989-6-15 12:00".
1.7.3 Conversion of the time
Some programs which work with NetCDF data can only process relative time axes. Therefore it may be necessary to convert from an absolute into a relative time axis. This conversion can be done for each operator with the CDO option ’-r’. To convert a relative into an absolute time axis use the CDO option ’-a’.
1.8 Parameter table
A parameter table is an ASCII formated file to convert code numbers to variable names. Each variable has one line with its code number, name and a description with optional units in a blank separated list. It can only be used for GRIB, SERVICE, EXTRA and IEG formated files. The CDO option ’-t <partab>’ sets the default parameter table for all input files. Use the operator ’setpartab’ to set the parameter table for a specific file.
Example of a CDO parameter table:
134 aps surface pressure [Pa] 141 sn snow depth [m] 147 ahfl latent heat flux [W/m**2] 172 slm land sea mask 175 albedo surface albedo 211 siced ice depth [m]
1.9 Missing values
Missing values are data points that are missing or invalid. Such data points are treated in a different way than valid data. Most CDO operators can handle missing values in a smart way. But if the missing value is within the range of valid data, it can lead to incorrect results. This applies to all arithmetic operations, but especially to logical operations when the missing value is 0 or 1.
The default missing value for GRIB, SERVICE, EXTRA and IEG files is -9.e33. The CDO option ’-m <missval>’ overwrites the default missing value. In NetCDF files the variable attribute ’_FillValue’ is used as a missing value. The operator ’setmissval’ can be used to set a new missing value.
The CDO use of the missing value is shown in the following tables, where one table is printed for each operation. The operations are applied to arbitrary numbers a, b, the special case 0, and the missing value miss. For example the table named "addition" shows that the sum of an arbitrary number a and the missing value is the missing value, and the table named "multiplication" shows that 0 multiplied by missing value results in 0.
| addition | b | miss | |
| a | a + b | miss | |
| miss | miss | miss | |
| subtraction | b | miss | |
| a | a - b | miss | |
| miss | miss | miss | |
| multiplication | b | 0 | miss |
| a | a * b | 0 | miss |
| 0 | 0 | 0 | 0 |
| miss | miss | 0 | miss |
| division | b | 0 | miss |
| a | a∕b | miss | miss |
| 0 | 0 | miss | miss |
| miss | miss | miss | miss |
| maximum | b | miss | |
| a | max(a,b) | a | |
| miss | b | miss | |
| minimum | b | miss | |
| a | min(a,b) | a | |
| miss | b | miss | |
| sum | b | miss | |
| a | a + b | a | |
| miss | b | miss | |
The handling of missing values by the operations "minimum" and "maximum" may be surprising, but the definition given here is more consistent with that expected in practice. Mathematical functions (e.g. log, sqrt, etc.) return the missing value if an argument is the missing value or an argument is out of range.
All statistical functions ignore missing values, treading them as not belonging to the sample, with the side-effect of a reduced sample size.
1.9.1 Mean and average
An artificial distinction is made between the notions mean and average. The mean is regarded as a statistical function, whereas the average is found simply by adding the sample members and dividing the result by the sample size. For example, the mean of 1, 2, miss and 3 is (1 + 2 + 3)∕3 = 2, whereas the average is (1 + 2 + miss + 3)∕4 = miss∕4 = miss. If there are no missing values in the sample, the average and mean are identical.
1.10 Percentile
There is no standard definition of percentile. All definitions yield to similar results when the number of values is very large. The following percentile methods are available in CDO:
| Percentile | |
| method |
Description |
| nrank | Nearest Rank method [default in CDO] |
| nist | The primary method recommended by NIST |
| rtype8 | R’s type=8 method |
| inverted_cdf | NumPy with percentile method=’inverted_cdf’ (R type=1) |
| averaged_inverted_cdf | NumPy with percentile method=’averaged_inverted_cdf’ (R type=2) |
| closest_observation | NumPy with percentile method=’closest_observation’ (R type=3) |
| interpolated_inverted_cdf | NumPy with percentile method=’interpolated_inverted_cdf’ (R type=4) |
| hazen | NumPy with percentile method=’hazen’ (R type=5) |
| weibull | NumPy with percentile method=’weibull’ (R type=6) |
| linear | NumPy with percentile method=’linear’ (R type=7) [default in NumPy and R] |
| median_unbiased | NumPy with percentile method=’median_unbiased’ (R type=8) |
| normal_unbiased | NumPy with percentile method=’normal_unbiased’ (R type=9) |
| lower | NumPy with percentile method=’lower’ |
| higher | NumPy with percentile method=’higher’ |
| midpoint | NumPy with percentile method=’midpoint’ |
| nearest | NumPy with percentile method=’nearest’ |
The percentile method can be selected with the CDO option --percentile. The Nearest Rank method is the default percentile method in CDO.
The different percentile methods can lead to different results, especially for small number of data values. Consider the ordered list {15, 20, 35, 40, 50, 55}, which contains six data values. Here is the result for the 30th, 40th, 50th, 75th and 100th percentiles of this list using the different percentile methods:
| Percentile | NumPy | NumPy | NumPy | NumPy | |||
| P | nrank | nist | rtype8 | linear | lower | higher | nearest |
| 30th | 20 | 21.5 | 23.5 | 27.5 | 20 | 35 | 35 |
| 40th | 35 | 32 | 33 | 35 | 35 | 35 | 35 |
| 50th | 35 | 37.5 | 37.5 | 37.5 | 35 | 40 | 40 |
| 75th | 50 | 51.25 | 50.42 | 47.5 | 40 | 50 | 50 |
| 100th | 55 | 55 | 55 | 55 | 55 | 55 | 55 |
1.10.1 Percentile over timesteps
The amount of data for time series can be very large. All data values need to held in memory to calculate the percentile. The percentile over timesteps uses a histogram algorithm, to limit the amount of required memory. The default number of histogram bins is 101. That means the histogram algorithm is used, when the dataset has more than 101 time steps. The default can be overridden by setting the environment variable CDO_PCTL_NBINS to a different value. The histogram algorithm is implemented only for the Nearest Rank method.
1.11 Regions
The CDO operators maskregion and selregion can be used to mask and select regions. For this purpose, the region needs to be defined by the user. In CDO there are two possibilities to define regions.
One possibility is to define the regions with an ASCII file. Each region is defined by a polygon. Each line of the polygon contains the longitude and latitude coordinates of a point. A description file for regions can contain several polygons, these must be separated by a line with the character &.
Here is a simple example of a polygon for a box with longitudes from 120W to 90E and latitudes from 20N to 20S:
120 20 120 -20 270 -20 270 20
With the second option, predefined regions can be used via country codes. A country is specified with dcw:<CountryCode>. Country codes can be combined with the plus sign.
The ISO two-letter country codes can be found on https://en.wikipedia.org/wiki/ISO_3166-1_alpha-2. To define a state, append the state code to the country code, e.g. USAK for Alaska. For the coordinates of a country CDO uses the DCW (Digital Chart of the World) dataset from GMT. This dataset must be installed on the system and the environment variable DIR_DCW must point to it.
2 Reference manual
This section gives a description of all operators. Related operators are grouped to modules. For easier description all single input files are named infile or infile1, infile2, etc., and an arbitrary number of input files are named infiles. All output files are named outfile or outfile1, outfile2, etc. Further the following notion is introduced:
-
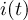
-
Timestep
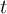 of infile
of infile
-
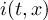
-
Element number
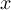 of the field at timestep
of the field at timestep 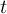 of infile
of infile
-
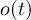
-
Timestep
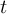 of outfile
of outfile
-
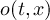
-
Element number
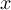 of the field at timestep
of the field at timestep 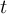 of outfile
of outfile
2.1 Information
This section contains modules to print information about datasets. All operators print there results to standard output.
Here is a short overview of all operators in this section:
| info | Dataset information listed by parameter identifier |
| infon | Dataset information listed by parameter name |
| cinfo | Compact information listed by parameter name |
| map | Dataset information and simple map |
| sinfo | Short information listed by parameter identifier |
| sinfon | Short information listed by parameter name |
| xsinfo | Extra short information listed by parameter name |
| xsinfop | Extra short information listed by parameter identifier |
| diff | Compare two datasets listed by parameter id |
| diffn | Compare two datasets listed by parameter name |
| npar | Number of parameters |
| nlevel | Number of levels |
| nyear | Number of years |
| nmon | Number of months |
| ndate | Number of dates |
| ntime | Number of timesteps |
| ngridpoints | Number of gridpoints |
| ngrids | Number of horizontal grids |
| showformat | Show file format |
| showcode | Show code numbers |
| showname | Show variable names |
| showstdname | Show standard names |
| showlevel | Show levels |
| showltype | Show GRIB level types |
| showyear | Show years |
| showmon | Show months |
| showdate | Show date information |
| showtime | Show time information |
| showtimestamp | Show timestamp |
| showchunkspec | Show chunk specification |
| showfilter | Show filter specification |
| showattribute | Show a global attribute or a variable attribute |
| partab | Parameter table |
| codetab | Parameter code table |
| griddes | Grid description |
| zaxisdes | Z-axis description |
| vct | Vertical coordinate table |
2.1.1 INFO - Information and simple statistics
Synopsis
<operator> infiles
Description
This module writes information about the structure and contents for each field of all input files to standard output. A field is a horizontal layer of a data variable. All input files need to have the same structure with the same variables on different timesteps. The information displayed depends on the chosen operator.
Operators
- info
-
Dataset information listed by parameter identifier
Prints information and simple statistics for each field of all input datasets. For each field the operator prints one line with the following elements:-
Date and Time
-
Level, Gridsize and number of Missing values
-
Minimum, Mean and Maximum
The mean value is computed without the use of area weights! -
Parameter identifier
-
- infon
-
Dataset information listed by parameter name
The same as operator info but using the name instead of the identifier to label the parameter. - cinfo
-
Compact information listed by parameter name
cinfo is a compact version of infon. It prints the minimum, mean and maximum value for each variable across all layers and time steps. - map
-
Dataset information and simple map
Prints information, simple statistics and a map for each field of all input datasets. The map will be printed only for fields on a regular lon/lat grid.
Example
To print information and simple statistics for each field of a dataset use:
cdo infon infile
This is an example result of a dataset with one 2D parameter over 12 timesteps:
-1 : Date Time Level Size Miss : Minimum Mean Maximum : Name 1 : 1987-01-31 12:00:00 0 2048 1361 : 232.77 266.65 305.31 : SST 2 : 1987-02-28 12:00:00 0 2048 1361 : 233.64 267.11 307.15 : SST 3 : 1987-03-31 12:00:00 0 2048 1361 : 225.31 267.52 307.67 : SST 4 : 1987-04-30 12:00:00 0 2048 1361 : 215.68 268.65 310.47 : SST 5 : 1987-05-31 12:00:00 0 2048 1361 : 215.78 271.53 312.49 : SST 6 : 1987-06-30 12:00:00 0 2048 1361 : 212.89 272.80 314.18 : SST 7 : 1987-07-31 12:00:00 0 2048 1361 : 209.52 274.29 316.34 : SST 8 : 1987-08-31 12:00:00 0 2048 1361 : 210.48 274.41 315.83 : SST 9 : 1987-09-30 12:00:00 0 2048 1361 : 210.48 272.37 312.86 : SST 10 : 1987-10-31 12:00:00 0 2048 1361 : 219.46 270.53 309.51 : SST 11 : 1987-11-30 12:00:00 0 2048 1361 : 230.98 269.85 308.61 : SST 12 : 1987-12-31 12:00:00 0 2048 1361 : 241.25 269.94 309.27 : SST
2.1.2 SINFO - Short information
Synopsis
<operator> infiles
Description
This module writes information about the structure of infiles to standard output. infiles is an arbitrary number of input files. All input files need to have the same structure with the same variables on different timesteps. The information displayed depends on the chosen operator.
Operators
- sinfo
-
Short information listed by parameter identifier
Prints short information of a dataset. The information is divided into 4 sections. Section 1 prints one line per parameter with the following information:-
institute and source
-
time c=constant v=varying
-
type of statistical processing
-
number of levels and z-axis number
-
horizontal grid size and number
-
data type
-
parameter identifier
Section 2 and 3 gives a short overview of all grid and vertical coordinates. And the last section contains short information of the time coordinate.
-
- sinfon
-
Short information listed by parameter name
The same as operator sinfo but using the name instead of the identifier to label the parameter.
Example
To print short information of a dataset use:
cdo sinfon infile
This is the result of an ECHAM5 dataset with 3 parameter over 12 timesteps:
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Name 1 : MPIMET ECHAM5 c instant 1 1 2048 1 F32 : GEOSP 2 : MPIMET ECHAM5 v instant 4 2 2048 1 F32 : T 3 : MPIMET ECHAM5 v instant 1 1 2048 1 F32 : TSURF Grid coordinates : 1 : gaussian : points=2048 (64x32) F16 longitude : 0 to 354.375 by 5.625 degrees_east circular latitude : 85.7606 to -85.7606 degrees_north Vertical coordinates : 1 : surface : levels=1 2 : pressure : levels=4 level : 92500 to 20000 Pa Time coordinate : time : 12 steps YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss YYYY-MM-DD hh:mm:ss 1987-01-31 12:00:00 1987-02-28 12:00:00 1987-03-31 12:00:00 1987-04-30 12:00:00 1987-05-31 12:00:00 1987-06-30 12:00:00 1987-07-31 12:00:00 1987-08-31 12:00:00 1987-09-30 12:00:00 1987-10-31 12:00:00 1987-11-30 12:00:00 1987-12-31 12:00:00
2.1.3 XSINFO - Extra short information
Synopsis
<operator> infiles
Description
This module writes information about the structure of infiles to standard output. infiles is an arbitrary number of input files. All input files need to have the same structure with the same variables on different timesteps. The information displayed depends on the chosen operator.
Operators
- xsinfo
-
Extra short information listed by parameter name
Prints short information of a dataset. The information is divided into 4 sections. Section 1 prints one line per parameter with the following information:-
institute and source
-
time c=constant v=varying
-
type of statistical processing
-
number of levels and z-axis number
-
horizontal grid size and number
-
data type
-
memory type (float or double)
-
parameter name
Section 2 to 4 gives a short overview of all grid, vertical and time coordinates.
-
- xsinfop
-
Extra short information listed by parameter identifier
The same as operator xsinfo but using the identifier instead of the name to label the parameter.
Example
To print extra short information of a dataset use:
cdo xsinfo infile
This is the result of an ECHAM5 dataset with 3 parameter over 12 timesteps:
-1 : Institut Source T Steptype Levels Num Points Num Dtype Mtype : Name 1 : MPIMET ECHAM5 c instant 1 1 2048 1 F32 F32 : GEOSP 2 : MPIMET ECHAM5 v instant 4 2 2048 1 F32 F32 : T 3 : MPIMET ECHAM5 v instant 1 1 2048 1 F32 F32 : TSURF Grid coordinates : 1 : gaussian : points=2048 (64x32) F16 longitude: 0 to 354.375 by 5.625 degrees_east circular latitude: 85.7606 to -85.7606 degrees_north Vertical coordinates : 1 : surface : levels=1 2 : pressure : levels=4 level: 92500 to 20000 Pa Time coordinate : steps: 12 time: 1987-01-31T18:00:00 to 1987-12-31T18:00:00 by 1 month units: days since 1987-01-01T00:00:00 calendar: proleptic_gregorian
2.1.4 DIFF - Compare two datasets field by field
Synopsis
<operator>[,parameter] infile1 infile2
Description
Compares the contents of two datasets field by field. The input datasets need to have the same structure and its fields need to have the dimensions. Try the option names if the number of variables differ. Exit status is 0 if inputs are the same and 1 if they differ.
Operators
- diff
-
Compare two datasets listed by parameter id
Provides statistics on differences between two datasets. For each pair of fields the operator prints one line with the following information:-
Date and Time
-
Level, Gridsize and number of Missing values
-
Number of different values
-
Occurrence of coefficient pairs with different signs (S)
-
Occurrence of zero values (Z)
-
Maxima of absolute difference of coefficient pairs
-
Maxima of relative difference of non-zero coefficient pairs with equal signs
-
Parameter identifier
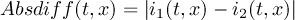
-
- diffn
-
Compare two datasets listed by parameter name
The same as operator diff. Using the name instead of the identifier to label the parameter.
Parameter
- maxcount
-
INTEGER Stop after maxcount different fields
- abslim
-
FLOAT Limit of the maximum absolute difference (default: 0)
- rellim
-
FLOAT Limit of the maximum relative difference (default: 1)
- names
-
STRING Consideration of the variable names of only one input file (left/right) or the intersection of both (intersect).
Example
To print the difference for each field of two datasets use:
cdo diffn infile1 infile2
This is an example result of two datasets with one 2D parameter over 12 timesteps:
Date Time Level Size Miss Diff : S Z Max_Absdiff Max_Reldiff : Name 1 : 1987-01-31 12:00:00 0 2048 1361 273 : F F 0.00010681 4.1660e-07 : SST 2 : 1987-02-28 12:00:00 0 2048 1361 309 : F F 6.1035e-05 2.3742e-07 : SST 3 : 1987-03-31 12:00:00 0 2048 1361 292 : F F 7.6294e-05 3.3784e-07 : SST 4 : 1987-04-30 12:00:00 0 2048 1361 183 : F F 7.6294e-05 3.5117e-07 : SST 5 : 1987-05-31 12:00:00 0 2048 1361 207 : F F 0.00010681 4.0307e-07 : SST 7 : 1987-07-31 12:00:00 0 2048 1361 317 : F F 9.1553e-05 3.5634e-07 : SST 8 : 1987-08-31 12:00:00 0 2048 1361 219 : F F 7.6294e-05 2.8849e-07 : SST 9 : 1987-09-30 12:00:00 0 2048 1361 188 : F F 7.6294e-05 3.6168e-07 : SST 10 : 1987-10-31 12:00:00 0 2048 1361 297 : F F 9.1553e-05 3.5001e-07 : SST 11 : 1987-11-30 12:00:00 0 2048 1361 234 : F F 6.1035e-05 2.3839e-07 : SST 12 : 1987-12-31 12:00:00 0 2048 1361 267 : F F 9.3553e-05 3.7624e-07 : SST 11 of 12 fields differ
2.1.5 NINFO - Print the number of parameters, levels or times
Synopsis
<operator> infile
Description
This module prints the number of variables, levels or times of the input dataset.
Operators
- npar
-
Number of parameters
Prints the number of parameters (variables). - nlevel
-
Number of levels
Prints the number of levels for each variable. - nyear
-
Number of years
Prints the number of different years. - nmon
-
Number of months
Prints the number of different combinations of years and months. - ndate
-
Number of dates
Prints the number of different dates. - ntime
-
Number of timesteps
Prints the number of timesteps. - ngridpoints
-
Number of gridpoints
Prints the number of gridpoints for each variable. - ngrids
-
Number of horizontal grids
Prints the number of horizontal grids.
Example
To print the number of parameters (variables) in a dataset use:
cdo npar infile
To print the number of months in a dataset use:
cdo nmon infile
2.1.6 SHOWINFO - Show variable information
Synopsis
<operator> infile
Description
This module prints meta-data information of all input variables. Depending on the chosen operator the name, level, date, time and other information is printed.
Operators
- showformat
-
Show file format
Prints the file format of the input dataset. - showcode
-
Show code numbers
Prints the code number of all variables. - showname
-
Show variable names
Prints the name of all variables. - showstdname
-
Show standard names
Prints the standard name of all variables. - showlevel
-
Show levels
Prints all levels for each variable. - showltype
-
Show GRIB level types
Prints the GRIB level type for all z-axes. - showyear
-
Show years
Prints all years. - showmon
-
Show months
Prints all months. - showdate
-
Show date information
Prints date information of all timesteps (format YYYY-MM-DD). - showtime
-
Show time information
Prints time information of all timesteps (format hh:mm:ss). - showtimestamp
-
Show timestamp
Prints timestamp of all timesteps (format YYYY-MM-DDThh:mm:ss). - showchunkspec
-
Show chunk specification
Prints NetCDF4 chunk specification of all variables. - showfilter
-
Show filter specification
Prints NetCDF4 filter specification of all variables.
Example
To print the code number of all variables in a dataset use:
cdo showcode infile
This is an example result of a dataset with three variables:
129 130 139
To print all months in a dataset use:
cdo showmon infile
This is an examples result of a dataset with an annual cycle:
1 2 3 4 5 6 7 8 9 10 11 12
2.1.7 SHOWATTRIBUTE - Show attributes
Synopsis
showattribute[,attributes] infile
Description
This operator prints the attributes of the data variables of a dataset.
Each attribute has the following structure:
[var_nm@][att_nm]
- var_nm
-
Variable name (optional). Example: pressure
- att_nm
-
Attribute name (optional). Example: units
The value of var_nm is the name of the variable containing the attribute (named att_nm) that you want to print. Use wildcards to print the attribute att_nm of more than one variable. A value of var_nm of ’*’ will print the attribute att_nm of all data variables. If var_nm is missing then att_nm refers to a global attribute.
The value of att_nm is the name of the attribute you want to print. Use wildcards to print more than one attribute. A value of att_nm of ’*’ will print all attributes.
Parameter
- attributes
-
STRING Comma-separated list of attributes.
2.1.8 FILEDES - Dataset description
Synopsis
<operator> infile
Description
This module provides operators to print meta information about a dataset. The printed meta-data depends on the chosen operator.
Operators
- partab
-
Parameter table
Prints all available meta information of the variables. - codetab
-
Parameter code table
Prints a code table with a description of all variables. For each variable the operator prints one line listing the code, name, description and units. - griddes
-
Grid description
Prints the description of all grids. - zaxisdes
-
Z-axis description
Prints the description of all z-axes. - vct
-
Vertical coordinate table
Prints the vertical coordinate table.
Example
Assume all variables of the dataset are on a regular Gausssian F16 grid. To print the grid description of this dataset use:
cdo griddes infile
Result:
gridtype : gaussian gridsize : 2048 xname : lon xlongname : longitude xunits : degrees_east yname : lat ylongname : latitude yunits : degrees_north xsize : 64 ysize : 32 xfirst : 0 xinc : 5.625 yvals : 85.76058 80.26877 74.74454 69.21297 63.67863 58.1429 52.6065 47.06964 41.53246 35.99507 30.4575 24.91992 19.38223 13.84448 8.306702 2.768903 -2.768903 -8.306702 -13.84448 -19.38223 -24.91992 -30.4575 -35.99507 -41.53246 -47.06964 -52.6065 -58.1429 -63.67863 -69.21297 -74.74454 -80.26877 -85.76058
2.2 File operations
This section contains modules to perform operations on files.
Here is a short overview of all operators in this section:
| apply | Apply operators on each input file. |
| copy | Copy datasets |
| clone | Clone datasets |
| cat | Concatenate datasets |
| tee | Duplicate a data stream |
| pack | Pack data |
| unpack | Unpack data |
| setchunkspec | Specify chunking |
| setfilter | Specify filter |
| bitrounding | Bit rounding |
| replace | Replace variables |
| duplicate | Duplicates a dataset |
| mergegrid | Merge grid |
| merge | Merge datasets with different fields |
| mergetime | Merge datasets sorted by date and time |
| splitcode | Split code numbers |
| splitparam | Split parameter identifiers |
| splitname | Split variable names |
| splitlevel | Split levels |
| splitgrid | Split grids |
| splitzaxis | Split z-axes |
| splittabnum | Split parameter table numbers |
| splithour | Split hours |
| splitday | Split days |
| splitseas | Split seasons |
| splityear | Split years |
| splityearmon | Split in years and months |
| splitmon | Split months |
| splitsel | Split time selection |
| splitdate | Splits a file into dates |
| distgrid | Distribute horizontal grid |
| collgrid | Collect horizontal grid |
2.2.1 APPLY - Apply operators
Synopsis
apply,operators infiles
Description
The apply utility runs the named operators on each input file. The input files must be enclosed in square brackets. This utility can only be used on a series of input files. These are all operators with more than one input file (infiles). Here is an incomplete list of these operators: copy, cat, merge, mergetime, select, ENSSTAT. The parameter operators is a blank-separated list of CDO operators. Use quotation marks if more than one operator is needed. Each operator may have only one input and output stream.
Parameter
- operators
-
STRING Blank-separated list of CDO operators.
Example
Suppose we have multiple input files with multiple variables on different time steps. The input files contain the variables U and V, among others. We are only interested in the absolute windspeed on all time steps. Here is the standard CDO solution for this task:
cdo expr,wind="sqrt(u*u+v*v)" -mergetime infile1 infile2 infile3 outfile
This first joins all the time steps together and then calculates the wind speed. If there are many variables in the input files, this procedure is ineffective. In this case it is better to first calculate the wind speed:
cdo mergetime -expr,wind="sqrt(u*u+v*v)" infile1 \ -expr,wind="sqrt(u*u+v*v)" infile2 \ -expr,wind="sqrt(u*u+v*v)" infile3 outfile
However, this can quickly become very confusing with more than 3 input files. The apply operator solves this problem:
cdo mergetime -apply,-expr,wind="sqrt(u*u+v*v)" [ infile1 infile2 infile3 ] outfile
Another example is the calculation of the mean value over several input files with ensmean. The input files contain several variables, but we are only interested in the variable named XXX:
cdo ensmean -apply,-selname,XXX [ infile1 infile2 infile3 ] outfile
2.2.2 COPY - Copy datasets
Synopsis
<operator> infiles outfile
Description
This module contains operators to copy, clone or concatenate datasets. infiles is an arbitrary number of input files. All input files need to have the same structure with the same variables on different timesteps.
Operators
- copy
-
Copy datasets
Copies all input datasets to outfile. - clone
-
Clone datasets
Copies all input datasets to outfile. In contrast to the copy operator, clone tries not to change the input data. GRIB records are neither decoded nor decompressed. - cat
-
Concatenate datasets
Concatenates all input datasets and appends the result to the end of outfile. If outfile does not exist it will be created.
Example
To change the format of a dataset to NetCDF use:
cdo -f nc copy infile outfile.nc
Add the option ’-r’ to create a relative time axis, as is required for proper recognition by GrADS or Ferret:
cdo -r -f nc copy infile outfile.nc
To concatenate 3 datasets with different timesteps of the same variables use:
cdo copy infile1 infile2 infile3 outfile
If the output dataset already exists and you wish to extend it with more timesteps use:
cdo cat infile1 infile2 infile3 outfile
2.2.3 TEE - Duplicate a data stream and write it to file
Synopsis
tee,outfile2 infile outfile1
Description
This operator copies the input dataset to outfile1 and outfile2. The first output stream in outfile1 can be further processesd with other cdo operators. The second output outfile2 is written to disk. It can be used to store intermediate results to a file.
Parameter
- outfile2
-
STRING Destination filename for the copy of the input file
Example
To compute the daily and monthy average of a dataset use:
cdo monavg -tee,outfile_dayavg dayavg infile outfile_monavg
2.2.4 PACK - Pack data
Synopsis
pack[,parameter] infile outfile
Description
Packing reduces the data volume by reducing the precision of the stored numbers. It is implemented using the NetCDF attributes add_offset and scale_factor. The operator pack calculates the attributes add_offset and scale_factor for all variables. The default data type for all variables is automatically changed to 16-bit integer. Use the CDO option -b to change the data type to a different integer precision, if needed. Missing values are automatically transformed to the current data type.
Alternatively, the pack parameters add_offset and scale_factor can be read from a file for each variable.
Parameter
- printparam
-
BOOL Print pack parameters to stdout for each variable
- filename
-
STRING Read pack parameters from file for each variable[format: name=<> add_offset=<> scale_factor=<>]
2.2.5 UNPACK - Unpack data
Synopsis
unpack infile outfile
Description
Packing reduces the data volume by reducing the precision of the stored numbers. It is implemented using the NetCDF attributes add_offset and scale_factor. The operator unpack unpack all packed variables. The default data type for all variables is automatically changed to 32-bit floats. Use the CDO option -b F64 to change the data type to 64-bit floats, if needed.
2.2.6 SETCHUNKSPEC - Specify chunking
Synopsis
setchunkspec,parameter infile outfile
Description
Specify chunking for selected variables in the output. Chunking is available for NetCDF4 and useful to specify the units of disk access and compression. The filename parameter is used to specify the file which contains the chunk specification for each variable. The chunkspec argument is a comma-separated string with the chunk size for the dimensions x,y,z,t. A chunkspec must name at least one dimension, e.g. t=<chunksize> to set the chunk size of the time dimension to <chunksize>.
Use the CDO option --chunkspec instead of setchunkspec if all variables require the same chunks.
Parameter
- filename
-
STRING Name of the file containing the chunk specification per variable [format: varname="<chunkspec>"]
2.2.7 SETFILTER - Specify filter
Synopsis
setfilter,parameter infile outfile
Description
Specify filter for selected variables in the output. Filters are available for NetCDF4 and mainly used to compress/decompress data. NetCDF4 uses the HDF5 plugins for filter support. To find the HDF5 plugins, the environment variable HDF5_PLUGIN_PATH must point to the directory with the installed plugins. The program may terminate unexpectedly if filters are used whose plugins are not found.
The filename parameter is used to specify the file which contains the filter specification for each variable. A filter specification consists of the filterId and the filter parameters. CDO supports multiple filters connected with ’|’. Here is a filter specification for bzip2 (filterId: 307) combined with szip (filterId:4): "307,9|4,32,32".
Use the CDO option --filter instead of setfilter if all variables require the same filter. More information about NetCDF4 filters can be found in https://docs.unidata.ucar.edu/netcdf-c/current/filters.html.
Parameter
- filename
-
STRING Name of the file containing the filter specification per variable [format: varname="<filterspec>"]
2.2.8 BITROUNDING - Bit rounding
Synopsis
bitrounding[,parameter] infile outfile
Description
This operator calculates for each field the number of necessary mantissa bits to get a certain information level in the data. With this number of significant bits (numbits) a rounding of the data is performed. This allows the data to be compressed to a higher level.
The default value of the information level is 0.9999 and can be adjusted with the parameter inflevel. That means 99.99% of the information in the mantissa bits is preserved.
Alternatively, the number of significant bits can be set for all variables with the numbits parameter. Furthermore, numbits can be assigned for each variable via the filename parameter. In this case, numbits is still calculated for all variables if they are not present in the file.
The analysis of the bit information is based on the Julia library BitInformation.jl. The procedure to derive the number of significant mantissa bits was adapted from the Python library xbitinfo. Quantize to the number of mantissa bits is done with IEEE rounding using code from NetCDF 4.9.0.
Currently only 32-bit float data is rounded. Data with missing values are not yet supported for the calculation of significant bits.
Parameter
- inflevel
-
FLOAT Information level (0 - 1) [default: 0.9999]
- addbits
-
INTEGER Add bits to the number of significant bits [default: 0]
- minbits
-
INTEGER Minimum value of the number of bits [default: 1]
- maxbits
-
INTEGER Maximum value of the number of bits [default: 23]
- numsteps
-
INTEGER Set to 1 to run the calculation only in the first time step
- numbits
-
INTEGER Set number of significant bits
- printbits
-
BOOL Print max. numbits per variable of 1st timestep to stdout [format: name=numbits]
- filename
-
STRING Read number of significant bits per variable from file [format: name=numbits]
Example
Apply bit rounding to all 32-bit float fields, preserving 99.9% of the information, followed by compression and storage to NetCDF4:
cdo -f nc4 -z zip bitrounding,inflevel=0.999 infile outfile
Add the option ’-v’ to view used number of mantissa bits for each field:
cdo -v -f nc4 -z zip bitrounding,inflevel=0.999 infile outfile
2.2.9 REPLACE - Replace variables
Synopsis
replace infile1 infile2 outfile
Description
This operator replaces variables in infile1 by variables from infile2 and write the result to outfile. Both input datasets need to have the same number of timesteps. All variable names may only occur once!
Example
Assume the first input dataset infile1 has three variables with the names geosp, t and tslm1 and the second input dataset infile2 has only the variable tslm1. To replace the variable tslm1 in infile1 by tslm1 from infile2 use:
cdo replace infile1 infile2 outfile
2.2.10 DUPLICATE - Duplicates a dataset
Synopsis
duplicate[,ndup] infile outfile
Description
This operator duplicates the contents of infile and writes the result to outfile. The optional parameter sets the number of duplicates, the default is 2.
Parameter
- ndup
-
INTEGER Number of duplicates, default is 2.
2.2.11 MERGEGRID - Merge grid
Synopsis
mergegrid infile1 infile2 outfile
Description
Merges grid points of all variables from infile2 to infile1 and write the result to outfile. Only the non missing values of infile2 will be used. The horizontal grid of infile2 should be smaller or equal to the grid of infile1 and the resolution must be the same. Only rectilinear grids are supported. Both input files need to have the same variables and the same number of timesteps.
2.2.12 MERGE - Merge datasets
Synopsis
merge infiles outfile
mergetime[,options] infiles outfile
Description
This module reads datasets from several input files, merges them and writes the resulting dataset to outfile.
Operators
- merge
-
Merge datasets with different fields
Merges time series of different fields from several input datasets. The number of fields per timestep written to outfile is the sum of the field numbers per timestep in all input datasets. The time series on all input datasets are required to have different fields and the same number of timesteps. The fields in each different input file either have to be different variables or different levels of the same variable. A mixture of different variables on different levels in different input files is not allowed. - mergetime
-
Merge datasets sorted by date and time
Merges all timesteps of all input files sorted by date and time. All input files need to have the same structure with the same variables on different timesteps. After this operation every input timestep is in outfile and all timesteps are sorted by date and time.
Parameter
- skip_same_time
-
BOOL Skips all consecutive timesteps with a double entry of the same timestamp.
- names
-
STRING Fill missing variable names with missing values (union) or use the intersection (intersect).
Note
Operators of this module need to open all input files simultaneously. The maximum number of open files depends on the operating system!
Example
Assume three datasets with the same number of timesteps and different variables in each dataset. To merge these datasets to a new dataset use:
cdo merge infile1 infile2 infile3 outfile
Assume you split a 6 hourly dataset with splithour. This produces four datasets, one for each hour. The following command merges them together:
cdo mergetime infile1 infile2 infile3 infile4 outfile
2.2.13 SPLIT - Split a dataset
Synopsis
<operator>[,parameter] infile obase
Description
This module splits infile into pieces. The output files will be named <obase><xxx><suffix> where suffix is the filename extension derived from the file format. xxx and the contents of the output files depends on the chosen operator. params is a comma-separated list of processing parameters.
Operators
- splitcode
-
Split code numbers
Splits a dataset into pieces, one for each different code number. xxx will have three digits with the code number. - splitparam
-
Split parameter identifiers
Splits a dataset into pieces, one for each different parameter identifier. xxx will be a string with the parameter identifier. - splitname
-
Split variable names
Splits a dataset into pieces, one for each variable name. xxx will be a string with the variable name. - splitlevel
-
Split levels
Splits a dataset into pieces, one for each different level. xxx will have six digits with the level. - splitgrid
-
Split grids
Splits a dataset into pieces, one for each different grid. xxx will have two digits with the grid number. - splitzaxis
-
Split z-axes
Splits a dataset into pieces, one for each different z-axis. xxx will have two digits with the z-axis number. - splittabnum
-
Split parameter table numbers
Splits a dataset into pieces, one for each GRIB1 parameter table number. xxx will have three digits with the GRIB1 parameter table number.
Parameter
- swap
-
STRING Swap the position of obase and xxx in the output filename
- uuid=<attname>
-
STRING Add a UUID as global attribute <attname> to each output file
Environment
- CDO_FILE_SUFFIX
-
Set the default file suffix. This suffix will be added to the output file names instead of the filename extension derived from the file format. Set this variable to NULL to disable the adding of a file suffix.
Note
Operators of this module need to open all output files simultaneously. The maximum number of open files depends on the operating system!
Example
Assume an input GRIB1 dataset with three variables, e.g. code number 129, 130 and 139. To split this dataset into three pieces, one for each code number use:
cdo splitcode infile code
Result of ’dir code*’:
code129.grb code130.grb code139.grb
2.2.14 SPLITTIME - Split timesteps of a dataset
Synopsis
<operator> infile obase
splitmon[,format] infile obase
Description
This module splits infile into timesteps pieces. The output files will be named <obase><xxx><suffix> where suffix is the filename extension derived from the file format. xxx and the contents of the output files depends on the chosen operator.
Operators
- splithour
-
Split hours
Splits a file into pieces, one for each different hour. xxx will have two digits with the hour. - splitday
-
Split days
Splits a file into pieces, one for each different day. xxx will have two digits with the day. - splitseas
-
Split seasons
Splits a file into pieces, one for each different season. xxx will have three characters with the season. - splityear
-
Split years
Splits a file into pieces, one for each different year. xxx will have four digits with the year (YYYY). - splityearmon
-
Split in years and months
Splits a file into pieces, one for each different year and month. xxx will have six digits with the year and month (YYYYMM). - splitmon
-
Split months
Splits a file into pieces, one for each different month. xxx will have two digits with the month.
Parameter
- format
-
STRING C-style format for strftime() (e.g. %B for the full month name)
Environment
- CDO_FILE_SUFFIX
-
Set the default file suffix. This suffix will be added to the output file names instead of the filename extension derived from the file format. Set this variable to NULL to disable the adding of a file suffix.
Note
Operators of this module need to open all output files simultaneously. The maximum number of open files depends on the operating system!
Example
Assume the input GRIB1 dataset has timesteps from January to December. To split each month with all variables into one separate file use:
cdo splitmon infile mon
Result of ’dir mon*’:
mon01.grb mon02.grb mon03.grb mon04.grb mon05.grb mon06.grb mon07.grb mon08.grb mon09.grb mon10.grb mon11.grb mon12.grb
2.2.15 SPLITSEL - Split selected timesteps
Synopsis
splitsel,nsets[,noffset[,nskip]] infile obase
Description
This operator splits infile into pieces, one for each adjacent sequence t1,....,t_n of timesteps of the same selected time range. The output files will be named <obase><nnnnnn><suffix> where nnnnnn is the sequence number and suffix is the filename extension derived from the file format.
Parameter
- nsets
-
INTEGER Number of input timesteps for each output file
- noffset
-
INTEGER Number of input timesteps skipped before the first timestep range (optional)
- nskip
-
INTEGER Number of input timesteps skipped between timestep ranges (optional)
Environment
- CDO_FILE_SUFFIX
-
Set the default file suffix. This suffix will be added to the output file names instead of the filename extension derived from the file format. Set this variable to NULL to disable the adding of a file suffix.
2.2.16 SPLITDATE - Splits a file into dates
Synopsis
splitdate infile obase
Description
This operator splits infile into pieces, one for each different date. The output files will be named <obase><YYYY-MM-DD><suffix> where YYYY-MM-DD is the date and suffix is the filename extension derived from the file format.
Environment
- CDO_FILE_SUFFIX
-
Set the default file suffix. This suffix will be added to the output file names instead of the filename extension derived from the file format. Set this variable to NULL to disable the adding of a file suffix.
2.2.17 DISTGRID - Distribute horizontal grid
Synopsis
distgrid,nx[,ny] infile obase
Description
This operator distributes a dataset into smaller pieces. Each output file contains a different region of the horizontal source grid. 2D Lon/Lat grids can be split into nx*ny pieces, where a target grid region contains a structured longitude/latitude box of the source grid. Data on an unstructured grid is split into nx pieces. The output files will be named <obase><xxx><suffix> where suffix is the filename extension derived from the file format. xxx will have five digits with the number of the target region.
Parameter
- nx
-
INTEGER Number of regions in x direction, or number of pieces for unstructured grids
- ny
-
INTEGER Number of regions in y direction [default: 1]
Note
This operator needs to open all output files simultaneously. The maximum number of open files depends on the operating system!
Example
Distribute data on a 2D Lon/Lat grid into 6 smaller files, each output file receives one half of x and a third of y of the source grid:
cdo distgrid,2,3 infile.nc obase
Below is a schematic illustration of this example:
On the left side is the data of the input file and on the right side is the data of the six output files.
2.2.18 COLLGRID - Collect horizontal grid
Synopsis
collgrid[,parameter] infiles outfile
Description
This operator collects the data of the input files to one output file. All input files need to have the same variables and the same number of timesteps on a different horizonal grid region. If the source regions are on a structured lon/lat grid, all regions together must result in a new structured lat/long grid box. Data on an unstructured grid are concatenated in the order of the input files. For ICON restart data, the array global_cell_indices is used for indexing if it is available. The parameter nx needs to be specified only for curvilinear grids.
Parameter
- nx
-
INTEGER Number of regions in x direction [default: number of input files]
- name
-
STRING Comma-separated list of variable names.
- levidx
-
INTEGER Comma-separated list or first/last[/inc] range of index of levels.
- gridtype
-
STRING For unstructured grids, set to unstructured.
Note
This operator needs to open all input files simultaneously. The maximum number of open files depends on the operating system!
Example
Collect the horizonal grid of 6 input files. Each input file contains a lon/lat region of the target grid:
cdo collgrid infile[1-6] outfile
Below is a schematic illustration of this example:
On the left side is the data of the six input files and on the right side is the collected data of the output file.
2.3 Selection
This section contains modules to select time steps, fields or a part of a field from a dataset.
Here is a short overview of all operators in this section:
| select | Select fields |
| delete | Delete fields |
| selmulti | Select multiple fields |
| delmulti | Delete multiple fields |
| changemulti | Change identication of multiple fields |
| selparam | Select parameters by identifier |
| delparam | Delete parameters by identifier |
| selcode | Select parameters by code number |
| delcode | Delete parameters by code number |
| selname | Select parameters by name |
| delname | Delete parameters by name |
| selstdname | Select parameters by standard name |
| sellevel | Select levels |
| sellevidx | Select levels by index |
| selgrid | Select grids |
| selzaxis | Select z-axes |
| selzaxisname | Select z-axes by name |
| selltype | Select GRIB level types |
| seltabnum | Select parameter table numbers |
| seltimestep | Select timesteps |
| seltime | Select times |
| selhour | Select hours |
| selday | Select days |
| selmonth | Select months |
| selyear | Select years |
| selseason | Select seasons |
| seldate | Select dates |
| selsmon | Select single month |
| sellonlatbox | Select a longitude/latitude box |
| selindexbox | Select an index box |
| selregion | Select cells inside regions |
| selcircle | Select cells inside a circle |
| selgridcell | Select grid cells |
| delgridcell | Delete grid cells |
| samplegrid | Resample grid |
| selyearidx | Select year by index |
| seltimeidx | Select timestep by index |
| bottomvalue | Extract bottom level |
| topvalue | Extract top level |
| isosurface | Extract isosurface |
2.3.1 SELECT - Select fields
Synopsis
<operator>,parameter infiles outfile
Description
This module selects some fields from infiles and writes them to outfile. infiles is an arbitrary number of input files. All input files need to have the same structure with the same variables on different timesteps. The fields selected depends on the chosen parameters. Parameter is a comma-separated list of "key=value" pairs. A range of integer values can be specified by first/last[/inc]. Wildcards are supported for string values.
Operators
- select
-
Select fields
Selects all fields with parameters in a user given list. - delete
-
Delete fields
Deletes all fields with parameters in a user given list.
Parameter
- name
-
STRING Comma-separated list of variable names.
- param
-
STRING Comma-separated list of parameter identifiers.
- code
-
INTEGER Comma-separated list or first/last[/inc] range of code numbers.
- level
-
FLOAT Comma-separated list of vertical levels.
- levrange
-
FLOAT First and last value of the level range.
- levidx
-
INTEGER Comma-separated list or first/last[/inc] range of index of levels.
- zaxisname
-
STRING Comma-separated list of zaxis names.
- zaxisnum
-
INTEGER Comma-separated list or first/last[/inc] range of zaxis numbers.
- ltype
-
INTEGER Comma-separated list or first/last[/inc] range of GRIB level types.
- gridname
-
STRING Comma-separated list of grid names.
- gridnum
-
INTEGER Comma-separated list or first/last[/inc] range of grid numbers.
- steptype
-
STRING Comma-separated list of timestep types (constant|avg|accum|min|max|range|diff|sum)
- date
-
STRING Comma-separated list of dates (format: YYYY-MM-DDThh:mm:ss).
- startdate
-
STRING Start date (format: YYYY-MM-DDThh:mm:ss).
- enddate
-
STRING End date (format: YYYY-MM-DDThh:mm:ss).
- minute
-
INTEGER Comma-separated list or first/last[/inc] range of minutes.
- hour
-
INTEGER Comma-separated list or first/last[/inc] range of hours.
- day
-
INTEGER Comma-separated list or first/last[/inc] range of days.
- month
-
INTEGER Comma-separated list or first/last[/inc] range of months.
- season
-
STRING Comma-separated list of seasons (substring of DJFMAMJJASOND or ANN).
- year
-
INTEGER Comma-separated list or first/last[/inc] range of years.
- dom
-
STRING Comma-separated list of the day of month (e.g. 29feb).
- timestep
-
INTEGER Comma-separated list or first/last[/inc] range of timesteps. Negative values select timesteps from the end (NetCDF only).
- timestep_of_year
-
INTEGER Comma-separated list or first/last[/inc] range of timesteps of year.
- timestepmask
-
STRING Read timesteps from a mask file.
Example
Assume you have 3 inputfiles. Each inputfile contains the same variables for a different time period. To select the variable T,U and V on the levels 200, 500 and 850 from all 3 input files, use:
cdo select,name=T,U,V,level=200,500,850 infile1 infile2 infile3 outfile
To remove the February 29th use:
cdo delete,dom=29feb infile outfile
2.3.2 SELMULTI - Select multiple fields via GRIB1 parameters
Synopsis
<operator>,selection-specification infile outfile
Description
This module selects multiple fields from infile and writes them to outfile. selection-specification is a filename or in-place string with the selection specification. Each selection-specification has the following compact notation format:
<type>(parameters; leveltype(s); levels)
- type
-
sel for select or del for delete (optional)
- parameters
-
GRIB1 parameter code number
- leveltype
-
GRIB1 level type
- levels
-
value of each level
Examples:
(1; 103; 0) (33,34; 105; 10) (11,17; 105; 2) (71,73,74,75,61,62,65,117,67,122,121,11,131,66,84,111,112; 105; 0)
The following descriptive notation can also be used for selection specification from a file:
SELECT/DELETE, PARAMETER=parameters, LEVTYPE=leveltye(s), LEVEL=levels
Examples:
SELECT, PARAMETER=1, LEVTYPE=103, LEVEL=0 SELECT, PARAMETER=33/34, LEVTYPE=105, LEVEL=10 SELECT, PARAMETER=11/17, LEVTYPE=105, LEVEL=2 SELECT, PARAMETER=71/73/74/75/61/62/65/117/67/122, LEVTYPE=105, LEVEL=0 DELETE, PARAMETER=128, LEVTYPE=109, LEVEL=*
The following will convert Pressure from Pa into hPa; Temp from Kelvin to Celsius:
SELECT, PARAMETER=1, LEVTYPE= 103, LEVEL=0, SCALE=0.01 SELECT, PARAMETER=11, LEVTYPE=105, LEVEL=2, OFFSET=273.15
If SCALE and/or OFFSET are defined, then the data values are scaled as SCALE*(VALUE-OFFSET).
Operators
- selmulti
-
Select multiple fields
- delmulti
-
Delete multiple fields
- changemulti
-
Change identication of multiple fields
Example
Change ECMWF GRIB code of surface pressure to Hirlam notation:
cdo changemulti,’{(134;1;*|1;105;*)}’ infile outfile
2.3.3 SELVAR - Select fields
Synopsis
<operator>,parameter infile outfile
selcode,codes infile outfile
delcode,codes infile outfile
selname,names infile outfile
delname,names infile outfile
selstdname,stdnames infile outfile
sellevel,levels infile outfile
sellevidx,levidx infile outfile
selgrid,grids infile outfile
selzaxis,zaxes infile outfile
selzaxisname,zaxisnames infile outfile
selltype,ltypes infile outfile
seltabnum,tabnums infile outfile
Description
This module selects some fields from infile and writes them to outfile. The fields selected depends on the chosen operator and the parameters. A range of integer values can be specified by first/last[/inc].
Operators
- selparam
-
Select parameters by identifier
Selects all fields with parameter identifiers in a user given list. - delparam
-
Delete parameters by identifier
Deletes all fields with parameter identifiers in a user given list. - selcode
-
Select parameters by code number
Selects all fields with code numbers in a user given list or range. - delcode
-
Delete parameters by code number
Deletes all fields with code numbers in a user given list or range. - selname
-
Select parameters by name
Selects all fields with parameter names in a user given list. - delname
-
Delete parameters by name
Deletes all fields with parameter names in a user given list. - selstdname
-
Select parameters by standard name
Selects all fields with standard names in a user given list. - sellevel
-
Select levels
Selects all fields with levels in a user given list. - sellevidx
-
Select levels by index
Selects all fields with index of levels in a user given list or range. - selgrid
-
Select grids
Selects all fields with grids in a user given list. - selzaxis
-
Select z-axes
Selects all fields with z-axes in a user given list. - selzaxisname
-
Select z-axes by name
Selects all fields with z-axis names in a user given list. - selltype
-
Select GRIB level types
Selects all fields with GRIB level type in a user given list or range. - seltabnum
-
Select parameter table numbers
Selects all fields with parameter table numbers in a user given list or range.
Parameter
- parameter
-
STRING Comma-separated list of parameter identifiers.
- codes
-
INTEGER Comma-separated list or first/last[/inc] range of code numbers.
- names
-
STRING Comma-separated list of variable names.
- stdnames
-
STRING Comma-separated list of standard names.
- levels
-
FLOAT Comma-separated list of vertical levels.
- levidx
-
INTEGER Comma-separated list or first/last[/inc] range of index of levels.
- ltypes
-
INTEGER Comma-separated list or first/last[/inc] range of GRIB level types.
- grids
-
STRING Comma-separated list of grid names or numbers.
- zaxes
-
STRING Comma-separated list of z-axis types or numbers.
- zaxisnames
-
STRING Comma-separated list of z-axis names.
- tabnums
-
INTEGER Comma-separated list or range of parameter table numbers.
Example
Assume an input dataset has three variables with the code numbers 129, 130 and 139. To select the variables with the code number 129 and 139 use:
cdo selcode,129,139 infile outfile
You can also select the code number 129 and 139 by deleting the code number 130 with:
cdo delcode,130 infile outfile
2.3.4 SELTIME - Select timesteps
Synopsis
seltimestep,timesteps infile outfile
seltime,times infile outfile
selhour,hours infile outfile
selday,days infile outfile
selmonth,months infile outfile
selyear,years infile outfile
selseason,seasons infile outfile
seldate,startdate[,enddate] infile outfile
selsmon,month[,nts1[,nts2]] infile outfile
Description
This module selects user specified timesteps from infile and writes them to outfile. The timesteps selected depends on the chosen operator and the parameters. A range of integer values can be specified by first/last[/inc].
Operators
- seltimestep
-
Select timesteps
Selects all timesteps with a timestep in a user given list or range. - seltime
-
Select times
Selects all timesteps with a time in a user given list or range. - selhour
-
Select hours
Selects all timesteps with a hour in a user given list or range. - selday
-
Select days
Selects all timesteps with a day in a user given list or range. - selmonth
-
Select months
Selects all timesteps with a month in a user given list or range. - selyear
-
Select years
Selects all timesteps with a year in a user given list or range. - selseason
-
Select seasons
Selects all timesteps with a month of a season in a user given list. - seldate
-
Select dates
Selects all timesteps with a date in a user given range. - selsmon
-
Select single month
Selects a month and optional an arbitrary number of timesteps before and after this month.
Parameter
- timesteps
-
INTEGER Comma-separated list or first/last[/inc] range of timesteps. Negative values select timesteps from the end (NetCDF only).
- times
-
STRING Comma-separated list of times (format hh:mm:ss).
- hours
-
INTEGER Comma-separated list or first/last[/inc] range of hours.
- days
-
INTEGER Comma-separated list or first/last[/inc] range of days.
- months
-
INTEGER Comma-separated list or first/last[/inc] range of months.
- years
-
INTEGER Comma-separated list or first/last[/inc] range of years.
- seasons
-
STRING Comma-separated list of seasons (substring of DJFMAMJJASOND or ANN).
- startdate
-
STRING Start date (format: YYYY-MM-DDThh:mm:ss).
- enddate
-
STRING End date (format: YYYY-MM-DDThh:mm:ss) [default: startdate].
- nts1
-
INTEGER Number of timesteps before the selected month [default: 0].
- nts2
-
INTEGER Number of timesteps after the selected month [default: nts1].
2.3.5 SELBOX - Select a box
Synopsis
sellonlatbox,lon1,lon2,lat1,lat2 infile outfile
selindexbox,idx1,idx2,idy1,idy2 infile outfile
Description
Selects grid cells inside a lon/lat or index box.
Operators
- sellonlatbox
-
Select a longitude/latitude box
Selects grid cells inside a lon/lat box. The user must specify the longitude and latitude of the edges of the box. Only those grid cells are considered whose grid center lies within the lon/lat box. For rotated lon/lat grids the parameters must be specified in rotated coordinates. - selindexbox
-
Select an index box
Selects grid cells within an index box. The user must specify the indices of the edges of the box. The index of the left edge can be greater then the one of the right edge. Use negative indexing to start from the end. The input grid must be a regular lon/lat or a 2D curvilinear grid.
Parameter
- lon1
-
FLOAT Western longitude in degrees
- lon2
-
FLOAT Eastern longitude in degrees
- lat1
-
FLOAT Southern or northern latitude in degrees
- lat2
-
FLOAT Northern or southern latitude in degrees
- idx1
-
INTEGER Index of first longitude (1 - nlon)
- idx2
-
INTEGER Index of last longitude (1 - nlon)
- idy1
-
INTEGER Index of first latitude (1 - nlat)
- idy2
-
INTEGER Index of last latitude (1 - nlat)
Example
To select the region with the longitudes from 30W to 60E and latitudes from 30N to 80N from all input fields use:
cdo sellonlatbox,-30,60,30,80 infile outfile
If the input dataset has fields on a regular Gaussian F16 grid, the same box can be selected with selindexbox by:
cdo selindexbox,60,11,3,11 infile outfile
2.3.6 SELREGION - Select horizontal regions
Synopsis
selregion,regions infile outfile
selcircle[,parameter] infile outfile
Description
Selects all grid cells with the center point inside user defined regions or a circle. The resulting grid is unstructured.
Operators
- selregion
-
Select cells inside regions
Selects all grid cells with the center point inside the regions. Regions can be defined by the user via an ASCII file. Each region consists of the geographic coordinates of a polygon. Each line of a polygon description file contains the longitude and latitude of one point. Each polygon description file can contain one or more polygons separated by a line with the character &.Predefined regions of countries can be specified via the country codes. A country is specified with dcw:<CountryCode>. Country codes can be combined with the plus sign.
- selcircle
-
Select cells inside a circle
Selects all grid cells with the center point inside a circle. The circle is described by geographic coordinates of the center and the radius of the circle.
Parameter
- regions
-
STRING Comma-separated list of ASCII formatted files with different regions
- lon
-
FLOAT Longitude of the center of the circle in degrees, default lon=0.0
- lat
-
FLOAT Latitude of the center of the circle in degrees, default lat=0.0
- radius
-
STRING Radius of the circle, default radius=1deg (units: deg, rad, km, m)
Example
To select all grid cells of a country use the country code with data from the Digital Chart of the World. Here is an example for Spain with the country code ES:
cdo selregion,dcw:ES infile outfile
2.3.7 SELGRIDCELL - Select grid cells
Synopsis
<operator>,indices infile outfile
Description
The operator selects grid cells of all fields from infile. The user must specify the index of each grid cell. The resulting grid in outfile is unstructured.
Operators
- selgridcell
-
Select grid cells
- delgridcell
-
Delete grid cells
Parameter
- indices
-
INTEGER Comma-separated list or first/last[/inc] range of indices
2.3.8 SAMPLEGRID - Resample grid
Synopsis
samplegrid,factor infile outfile
Description
This is a special operator for resampling the horizontal grid. No interpolation takes place. Resample factor=2 means every second grid point is removed. Only rectilinear and curvilinear source grids are supported by this operator.
Parameter
- factor
-
INTEGER Resample factor, typically 2, which will half the resolution
2.3.9 SELYEARIDX - Select year by index
Synopsis
selyearidx infile1 infile2 outfile
Description
Selects field elements from infile2 according to a year index from infile1. The index of the year in infile1 should be the result of corresponding yearminidx or yearmaxidx operations, respectively.
2.3.10 SELTIMEIDX - Select timestep by index
Synopsis
seltimeidx infile1 infile2 outfile
Description
Selects field elements from infile2 according to a timestep index from infile1. The index of the timestep in infile1 should be the result of corresponding timminidx or timmaxidx operations, respectively.
2.3.11 SELSURFACE - Extract surface
Synopsis
<operator> infile outfile
isosurface,isovalue infile outfile
Description
This module computes a surface from all 3D variables. The result is a horizonal 2D field.
Operators
- bottomvalue
-
Extract bottom level
This operator selects the valid values at the bottom level. The NetCDF CF compliant attribute positive is used to determine where top and bottom are. If this attribute is missing, low values are bottom and high values are top. - topvalue
-
Extract top level
This operator selects the valid values at the top level. The NetCDF CF compliant attribute positive is used to determine where top and bottom are. If this attribute is missing, low values are bottom and high values are top. - isosurface
-
Extract isosurface
This operator computes an isosurface. The value of the isosurfce is specified by the parameter isovalue. The isosurface is calculated by linear interpolation between two layers.
Parameter
- isovalue
-
FLOAT Isosurface value
2.4 Conditional selection
This section contains modules to conditional select field elements. The fields in the first input file are handled as a mask. A value not equal to zero is treated as "true", zero is treated as "false".
Here is a short overview of all operators in this section:
| ifthen | If then |
| ifnotthen | If not then |
| ifthenelse | If then else |
| ifthenc | If then constant |
| ifnotthenc | If not then constant |
| reducegrid | Reduce input file variables to locations, where mask is non-zero. |
2.4.1 COND - Conditional select one field
Synopsis
<operator> infile1 infile2 outfile
Description
This module selects field elements from infile2 with respect to infile1 and writes them to outfile. The fields in infile1 are handled as a mask. A value not equal to zero is treated as "true", zero is treated as "false". The number of fields in infile1 has either to be the same as in infile2 or the same as in one timestep of infile2 or only one. The fields in outfile inherit the meta data from infile2.
Operators
- ifthen
-
If then
o(t,x) =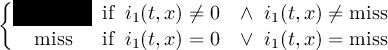
- ifnotthen
-
If not then
o(t,x) =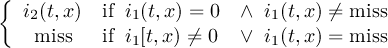
Example
To select all field elements of infile2 if the corresponding field element of infile1 is greater than 0 use:
cdo ifthen infile1 infile2 outfile
2.4.2 COND2 - Conditional select two fields
Synopsis
ifthenelse infile1 infile2 infile3 outfile
Description
This operator selects field elements from infile2 or infile3 with respect to infile1 and writes them to outfile. The fields in infile1 are handled as a mask. A value not equal to zero is treated as "true", zero is treated as "false". The number of fields in infile1 has either to be the same as in infile2 or the same as in one timestep of infile2 or only one. infile2 and infile3 need to have the same number of fields. The fields in outfile inherit the meta data from infile2.
o(t,x) = 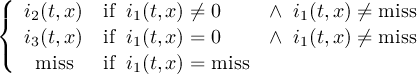
Example
To select all field elements of infile2 if the corresponding field element of infile1 is greater than 0 and from infile3 otherwise use:
cdo ifthenelse infile1 infile2 infile3 outfile
2.4.3 CONDC - Conditional select a constant
Synopsis
<operator>,c infile outfile
Description
This module creates fields with a constant value or missing value. The fields in infile are handled as a mask. A value not equal to zero is treated as "true", zero is treated as "false".
Operators
- ifthenc
-
If then constant
o(t,x) =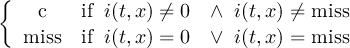
- ifnotthenc
-
If not then constant
o(t,x) =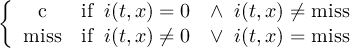
Parameter
- c
-
FLOAT Constant
Example
To create fields with the constant value 7 if the corresponding field element of infile is greater than 0 use:
cdo ifthenc,7 infile outfile
2.4.4 MAPREDUCE - Reduce fields to user-defined mask
Synopsis
reducegrid,mask[,limitCoordsOutput] infile outfile
Description
This module holds an operator for data reduction based on a user defined mask. The output grid is unstructured and includes coordinate bounds. Bounds can be avoided by using the additional ’nobounds’ keyword. With ’nocoords’ given, coordinates a completely suppressed.
Parameter
- mask
-
STRING file which holds the mask field
- limitCoordsOutput
-
STRING optional parameter to limit coordinates output: ’nobounds’ disables coordinate bounds, ’nocoords’ avoids all coordinate information
Example
To limit data fields to land values, a mask has to be created first with
cdo -gtc,0 -topo,ni96 lsm_gme96.grb
Here a GME grid is used. Say temp_gme96.grb contains a global temperture field. The following command limits the global grid to landpoints.
cdo -f nc reduce,lsm_gme96.grb temp_gme96.grb tempOnLand_gme96.nc
Note that output file type is NetCDF, because unstructured grids cannot be stored in GRIB format.
2.5 Comparison
This section contains modules to compare datasets. The resulting field is a mask containing 1 if the comparison is true and 0 if not.
Here is a short overview of all operators in this section:
| eq | Equal |
| ne | Not equal |
| le | Less equal |
| lt | Less than |
| ge | Greater equal |
| gt | Greater than |
| eqc | Equal constant |
| nec | Not equal constant |
| lec | Less equal constant |
| ltc | Less than constant |
| gec | Greater equal constant |
| gtc | Greater than constant |
| ymoneq | Compare time series with Equal |
| ymonne | Compare time series with NotEqual |
| ymonle | Compare time series with LessEqual |
| ymonlt | Compares if time series with LessThan |
| ymonge | Compares if time series with GreaterEqual |
| ymongt | Compares if time series with GreaterThan |
| yseaseq | Compare time series with Equal |
| yseasne | Compare time series with NotEqual |
| yseasle | Compare time series with LessEqual |
| yseaslt | Compares if time series with LessThan |
| yseasge | Compares if time series with GreaterEqual |
| yseasgt | Compares if time series with GreaterThan |
2.5.1 COMP - Comparison of two fields
Synopsis
<operator> infile1 infile2 outfile
Description
This module compares two datasets field by field. The resulting field is a mask containing 1 if the comparison is true and 0 if not. The number of fields in infile1 should be the same as in infile2. One of the input files can contain only one timestep or one field. The fields in outfile inherit the meta data from infile1 or infile2. The type of comparison depends on the chosen operator.
Operators
- eq
-
Equal
o(t,x) =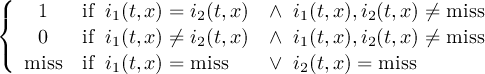
- ne
-
Not equal
o(t,x) =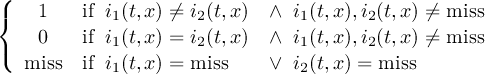
- le
-
Less equal
o(t,x) =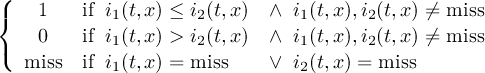
- lt
-
Less than
o(t,x) =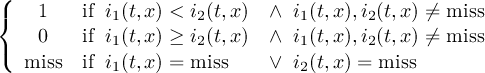
- ge
-
Greater equal
o(t,x) =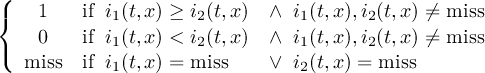
- gt
-
Greater than
o(t,x) =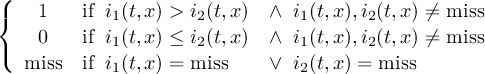
Example
To create a mask containing 1 if the elements of two fields are the same and 0 if the elements are different use:
cdo eq infile1 infile2 outfile
2.5.2 COMPC - Comparison of a field with a constant
Synopsis
<operator>,c infile outfile
Description
This module compares all fields of a dataset with a constant. The resulting field is a mask containing 1 if the comparison is true and 0 if not. The type of comparison depends on the chosen operator.
Operators
- eqc
-
Equal constant
o(t,x) =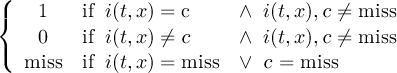
- nec
-
Not equal constant
o(t,x) =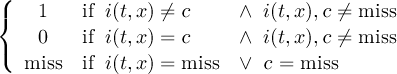
- lec
-
Less equal constant
o(t,x) =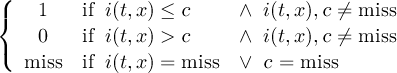
- ltc
-
Less than constant
o(t,x) =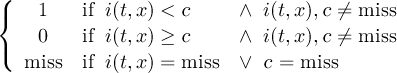
- gec
-
Greater equal constant
o(t,x) =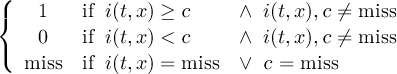
- gtc
-
Greater than constant
o(t,x) =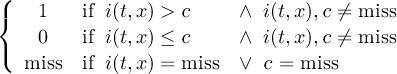
Parameter
- c
-
FLOAT Constant
Example
To create a mask containing 1 if the field element is greater than 273.15 and 0 if not use:
cdo gtc,273.15 infile outfile
2.5.3 YMONCOMP - Multi-year monthly comparison
Synopsis
<operator> infile1 infile2 outfile
Description
This module performs comparisons of a time series and one timestep with the same month of year. For each field in infile1 the corresponding field of the timestep in infile2 with the same month of year is used. The resulting field is a mask containing 1 if the comparison is true and 0 if not. The type of comparison depends on the chosen operator. The input files need to have the same structure with the same variables. Usually infile2 is generated by an operator of the module YMONSTAT.
Operators
- ymoneq
-
Compare time series with Equal
Compares whether a time series is equal to a multi-year monthly time series. - ymonne
-
Compare time series with NotEqual
Compares whether a time series is not equal to a multi-year monthly time series. - ymonle
-
Compare time series with LessEqual
Compares whether a time series is less than or equal to a multi-year monthly time series. - ymonlt
-
Compares if time series with LessThan
Compares whether a time series is less than a multi-year monthly time series. - ymonge
-
Compares if time series with GreaterEqual
Compares whether a time series is greater than or equal to a multi-year monthly time series. - ymongt
-
Compares if time series with GreaterThan
Compares whether a time series is greater than a multi-year monthly time series.
2.5.4 YSEASCOMP - Multi-year seasonal comparison
Synopsis
<operator> infile1 infile2 outfile
Description
This module performs comparisons of a time series and one timestep with the same season of year. For each field in infile1 the corresponding field of the timestep in infile2 with the same season of year is used. The resulting field is a mask containing 1 if the comparison is true and 0 if not. The type of comparison depends on the chosen operator. The input files need to have the same structure with the same variables. Usually infile2 is generated by an operator of the module YseasSTAT.
Operators
- yseaseq
-
Compare time series with Equal
Compares whether a time series is equal to a multi-year seasonal time series. - yseasne
-
Compare time series with NotEqual
Compares whether a time series is not equal to a multi-year seasonal time series. - yseasle
-
Compare time series with LessEqual
Compares whether a time series is less than or equal to a multi-year seasonal time series. - yseaslt
-
Compares if time series with LessThan
Compares whether a time series is less than a multi-year seasonal time series. - yseasge
-
Compares if time series with GreaterEqual
Compares whether a time series is greater than or equal to a multi-year seasonal time series. - yseasgt
-
Compares if time series with GreaterThan
Compares whether a time series is greater than a multi-year seasonal time series.
2.6 Modification
This section contains modules to modify the metadata, fields or part of a field in a dataset.
Here is a short overview of all operators in this section:
| setattribute | Set attributes |
| delattribute | Delete attributes |
| setpartabp | Set parameter table |
| setpartabn | Set parameter table |
| setcodetab | Set parameter code table |
| setcode | Set code number |
| setparam | Set parameter identifier |
| setname | Set variable name |
| setstdname | Set standard name |
| setunit | Set variable unit |
| setlevel | Set level |
| setltype | Set GRIB level type |
| setmaxsteps | Set max timesteps |
| setdate | Set date |
| settime | Set time of the day |
| setday | Set day |
| setmon | Set month |
| setyear | Set year |
| settunits | Set time units |
| settaxis | Set time axis |
| settbounds | Set time bounds |
| setreftime | Set reference time |
| setcalendar | Set calendar |
| shifttime | Shift timesteps |
| chcode | Change code number |
| chparam | Change parameter identifier |
| chname | Change variable or coordinate name |
| chunit | Change variable unit |
| chlevel | Change level |
| chlevelc | Change level of one code |
| chlevelv | Change level of one variable |
| setgrid | Set grid |
| setgridtype | Set grid type |
| setgridarea | Set grid cell area |
| setgridmask | Set grid mask |
| setprojparams | Set proj params |
| setzaxis | Set z-axis |
| genlevelbounds | Generate level bounds |
| invertlat | Invert latitudes |
| invertlev | Invert levels |
| shiftx | Shift x |
| shifty | Shift y |
| maskregion | Mask regions |
| masklonlatbox | Mask a longitude/latitude box |
| maskindexbox | Mask an index box |
| setclonlatbox | Set a longitude/latitude box to constant |
| setcindexbox | Set an index box to constant |
| enlarge | Enlarge fields |
| setmissval | Set a new missing value |
| setctomiss | Set constant to missing value |
| setmisstoc | Set missing value to constant |
| setrtomiss | Set range to missing value |
| setvrange | Set valid range |
| setmisstonn | Set missing value to nearest neighbor |
| setmisstodis | Set missing value to distance-weighted average |
| vertfillmiss | Vertical filling of missing values |
| timfillmiss | Temporal filling of missing values |
| setgridcell | Set the value of a grid cell |
2.6.1 SETATTRIBUTE - Set attributes
Synopsis
<operator>,attributes infile outfile
Description
This operator sets or deletes attributes of a dataset and writes the result to outfile. The new attributes are only available in outfile if the file format supports attributes.
Each attribute has the following structure:
[var_nm@]att_nm[:s|d|i]=[att_val|{[var_nm@]att_nm}]
- var_nm
-
Variable name (optional). Example: pressure
- att_nm
-
Attribute name. Example: units
- att_val
-
Comma-separated list of attribute values. Example: pascal
The value of var_nm is the name of the variable containing the attribute (named att_nm) that you want to set. Use wildcards to set the attribute att_nm to more than one variable. A value of var_nm of ’*’ will set the attribute att_nm to all data variables. If var_nm is missing then att_nm refers to a global attribute.
The value of att_nm is the name of the attribute you want to set. For each attribute a string (att_nm:s), a double (att_nm:d) or an integer (att_nm:i) type can be defined. By default the native type is set.
The value of att_val is the contents of the attribute att_nm. att_val may be a single value or one-dimensional array of elements. The type and the number of elements of an attribute will be detected automatically from the contents of the values. An already existing attribute att_nm will be overwritten or it will be removed if att_val is omitted. Alternatively, the values of an existing attribute can be copied. This attribute must then be enclosed in curly brackets.
A special meaning has the attribute name FILE. If this is the 1st attribute then all attributes are read from a file specified in the value of att_val.
Operators
- setattribute
-
Set attributes
- delattribute
-
Delete attributes
Parameter
- attributes
-
STRING Comma-separated list of attributes.
Note
Attributes are evaluated by CDO when opening infile. Therefor the result of this operator is not available for other operators when this operator is used in chaining operators.
Example
To set the units of the variable pressure to pascal use:
cdo setattribute,pressure@units=pascal infile outfile
To set the global text attribute "my_att" to "my contents", use:
cdo setattribute,my_att="my contents" infile outfile
Result of ’ncdump -h outfile’:
netcdf outfile { dimensions: ... variables: ... // global attributes: :my_att = "my contents" ; }
2.6.2 SETPARTAB - Set parameter table
Synopsis
<operator>,table[,convert] infile outfile
Description
This module transforms data and metadata of infile via a parameter table and writes the result to outfile. A parameter table is an ASCII formatted file with a set of parameter entries for each variable. Each new set have to start with "¶meter" and to end with "/".
The following parameter table entries are supported:
| Entry | Type | Description |
| name | WORD | Name of the variable |
| out_name | WORD | New name of the variable |
| param | WORD | Parameter identifier (GRIB1: code[.tabnum]; GRIB2: num[.cat[.dis]]) |
| out_param | WORD | New parameter identifier |
| type | WORD | Data type (real or double) |
| standard_name | WORD | As defined in the CF standard name table |
| long_name | STRING | Describing the variable |
| units | STRING | Specifying the units for the variable |
| comment | STRING | Information concerning the variable |
| cell_methods | STRING | Information concerning calculation of means or climatologies |
| cell_measures | STRING | Indicates the names of the variables containing cell areas and volumes |
| filterspec | STRING | NetCDF4 filter specification |
| missing_value | FLOAT | Specifying how missing data will be identified |
| valid_min | FLOAT | Minimum valid value |
| valid_max | FLOAT | Maximum valid value |
| ok_min_mean_abs | FLOAT | Minimum absolute mean |
| ok_max_mean_abs | FLOAT | Maximum absolute mean |
| factor | FLOAT | Scale factor |
| delete | INTEGER | Set to 1 to delete variable |
| convert | INTEGER | Set to 1 to convert the unit if necessary |
Unsupported parameter table entries are stored as variable attributes. The search key for the variable depends on the operator. Use setpartabn to search variables by the name. This is typically used for NetCDF datasets. The operator setpartabp searches variables by the parameter ID.
Operators
- setpartabp
-
Set parameter table
Search variables by the parameter identifier. - setpartabn
-
Set parameter table
Search variables by name.
Parameter
- table
-
STRING Parameter table file or name
- convert
-
STRING Converts the units if necessary
Example
Here is an example of a parameter table for one variable:
prompt> cat mypartab ¶meter name = t out_name = ta standard_name = air_temperature units = "K" missing_value = 1.0e+20 valid_min = 157.1 valid_max = 336.3 /
To apply this parameter table to a dataset use:
cdo setpartabn,mypartab,convert infile outfile
This command renames the variable t to ta. The standard name of this variable is set to air_temperature and the unit is set to [K] (converts the unit if necessary). The missing value will be set to 1.0e+20. In addition it will be checked whether the values of the variable are in the range of 157.1 to 336.3.
2.6.3 SET - Set field info
Synopsis
setcodetab,table infile outfile
setcode,code infile outfile
setparam,param infile outfile
setname,name infile outfile
setstdname,name infile outfile
setunit,unit infile outfile
setlevel,level infile outfile
setltype,ltype infile outfile
setmaxsteps,maxsteps infile outfile
Description
This module sets some field information. Depending on the chosen operator the parameter table, code number, parameter identifier, variable name or level is set.
Operators
- setcodetab
-
Set parameter code table
Sets the parameter code table for all variables. - setcode
-
Set code number
Sets the code number for all variables to the same given value. - setparam
-
Set parameter identifier
Sets the parameter identifier of the first variable. - setname
-
Set variable name
Sets the name of the first variable. - setstdname
-
Set standard name
Sets the standard name of the first variable. - setunit
-
Set variable unit
Sets the unit of the first variable. - setlevel
-
Set level
Sets the first level of all variables. - setltype
-
Set GRIB level type
Sets the GRIB level type of all variables. - setmaxsteps
-
Set max timesteps
Sets maximum number of timesteps
Parameter
- table
-
STRING Parameter table file or name
- code
-
INTEGER Code number
- param
-
STRING Parameter identifier (GRIB1: code[.tabnum]; GRIB2: num[.cat[.dis]])
- name
-
STRING Variable name
- level
-
FLOAT New level
- ltype
-
INTEGER GRIB level type
- maxsteps
-
INTEGER Maximum number of timesteps
2.6.4 SETTIME - Set time
Synopsis
setdate,date infile outfile
settime,time infile outfile
setday,day infile outfile
setmon,month infile outfile
setyear,year infile outfile
settunits,units infile outfile
settaxis,date,time[,inc] infile outfile
settbounds,frequency infile outfile
setreftime,date,time[,units] infile outfile
setcalendar,calendar infile outfile
shifttime,shiftValue infile outfile
Description
This module sets the time axis or part of the time axis. Which part of the time axis is overwritten/created depends on the chosen operator. The number of time steps does not change.
Operators
- setdate
-
Set date
Sets the date in every timestep to the same given value. - settime
-
Set time of the day
Sets the time in every timestep to the same given value. - setday
-
Set day
Sets the day in every timestep to the same given value. - setmon
-
Set month
Sets the month in every timestep to the same given value. - setyear
-
Set year
Sets the year in every timestep to the same given value. - settunits
-
Set time units
Sets the base units of a relative time axis. - settaxis
-
Set time axis
Sets the time axis. - settbounds
-
Set time bounds
Sets the time bounds. - setreftime
-
Set reference time
Sets the reference time of a relative time axis. - setcalendar
-
Set calendar
Sets the calendar attribute of a relative time axis. - shifttime
-
Shift timesteps
Shifts all timesteps by the parameter shiftValue.
Parameter
- day
-
INTEGER Value of the new day
- month
-
INTEGER Value of the new month
- year
-
INTEGER Value of the new year
- units
-
STRING Base units of the time axis (seconds|minutes|hours|days|months|years)
- date
-
STRING Date (format: YYYY-MM-DD)
- time
-
STRING Time (format: hh:mm:ss)
- inc
-
STRING Optional increment (seconds|minutes|hours|days|months|years) [default: 1hour]
- frequency
-
STRING Frequency of the time series (hour|day|month|year)
- calendar
-
STRING Calendar (standard|proleptic_gregorian|360_day|365_day|366_day)
- shiftValue
-
STRING Shift value (e.g. -3hour)
Example
To set the time axis to 1987-01-16 12:00:00 with an increment of one month for each timestep use:
cdo settaxis,1987-01-16,12:00:00,1mon infile outfile
Result of ’cdo showdate outfile’ for a dataset with 12 timesteps:
1987-01-16 1987-02-16 1987-03-16 1987-04-16 1987-05-16 1987-06-16 \ 1987-07-16 1987-08-16 1987-09-16 1987-10-16 1987-11-16 1987-12-16
To shift this time axis by -15 days use:
cdo shifttime,-15days infile outfile
Result of ’cdo showdate outfile’:
1987-01-01 1987-02-01 1987-03-01 1987-04-01 1987-05-01 1987-06-01 \ 1987-07-01 1987-08-01 1987-09-01 1987-10-01 1987-11-01 1987-12-01
2.6.5 CHANGE - Change field header
Synopsis
chcode,oldcode,newcode[,...] infile outfile
chparam,oldparam,newparam,... infile outfile
chname,oldname,newname,... infile outfile
chunit,oldunit,newunit,... infile outfile
chlevel,oldlev,newlev,... infile outfile
chlevelc,code,oldlev,newlev infile outfile
chlevelv,name,oldlev,newlev infile outfile
Description
This module reads fields from infile, changes some header values and writes the results to outfile. The kind of changes depends on the chosen operator.
Operators
- chcode
-
Change code number
Changes some user given code numbers to new user given values. - chparam
-
Change parameter identifier
Changes some user given parameter identifiers to new user given values. - chname
-
Change variable or coordinate name
Changes some user given variable or coordinate names to new user given names. - chunit
-
Change variable unit
Changes some user given variable units to new user given units. - chlevel
-
Change level
Changes some user given levels to new user given values. - chlevelc
-
Change level of one code
Changes one level of a user given code number. - chlevelv
-
Change level of one variable
Changes one level of a user given variable name.
Parameter
- code
-
INTEGER Code number
- oldcode,newcode,...
-
INTEGER Pairs of old and new code numbers
- oldparam,newparam,...
-
STRING Pairs of old and new parameter identifiers
- name
-
STRING Variable name
- oldname,newname,...
-
STRING Pairs of old and new variable names
- oldlev
-
FLOAT Old level
- newlev
-
FLOAT New level
- oldlev,newlev,...
-
FLOAT Pairs of old and new levels
Example
To change the code number 98 to 179 and 99 to 211 use:
cdo chcode,98,179,99,211 infile outfile
2.6.6 SETGRID - Set grid information
Synopsis
setgrid,grid infile outfile
setgridtype,gridtype infile outfile
setgridarea,gridarea infile outfile
setgridmask,gridmask infile outfile
setprojparams,projparams infile outfile
Description
This module modifies the metadata of the horizontal grid. Depending on the chosen operator a new grid description is set, the coordinates are converted or the grid cell area is added.
Operators
- setgrid
-
Set grid
Sets a new grid description. The input fields need to have the same grid size as the size of the target grid description. - setgridtype
-
Set grid type
Sets the grid type of all input fields. The following grid types are available:- curvilinear
-
Converts a regular grid to a curvilinear grid
- unstructured
-
Converts a regular or curvilinear grid to an unstructured grid
- dereference
-
Dereference a reference to a grid
- regular
-
Linear interpolation of a reduced Gaussian grid to a regular Gaussian grid
- regularnn
-
Nearest neighbor interpolation of a reduced Gaussian grid to a regular Gaussian grid
- lonlat
-
Converts a regular lonlat grid stored as a curvilinear grid back to a lonlat grid
- projection
-
Removes the geographical coordinates if projection parameter available
- setgridarea
-
Set grid cell area
Sets the grid cell area. The parameter gridarea is the path to a data file, the first field is used as grid cell area. The input fields need to have the same grid size as the grid cell area. The grid cell area is used to compute the weights of each grid cell if needed by an operator, e.g. for fldmean. - setgridmask
-
Set grid mask
Sets the grid mask. The parameter gridmask is the path to a data file, the first field is used as the grid mask. The input fields need to have the same grid size as the grid mask. The grid mask is used as the target grid mask for remapping, e.g. for remapbil. - setprojparams
-
Set proj params
Sets the proj_params attribute of a projection. This attribute is used to compute geographic coordinates of a projecton with the proj library.
Parameter
- grid
-
STRING Grid description file or name
- gridtype
-
STRING Grid type (curvilinear, unstructured, regular, lonlat, projection or dereference)
- gridarea
-
STRING Data file, the first field is used as grid cell area
- gridmask
-
STRING Data file, the first field is used as grid mask
- projparams
-
STRING Proj library parameter (e.g.:+init=EPSG:3413)
Example
Assuming a dataset has fields on a grid with 2048 elements without or with wrong grid description. To set the grid description of all input fields to a regular Gaussian F32 grid (8192 gridpoints) use:
cdo setgrid,F32 infile outfile
2.6.7 SETZAXIS - Set z-axis information
Synopsis
setzaxis,zaxis infile outfile
genlevelbounds[,zbot[,ztop]] infile outfile
Description
This module modifies the metadata of the vertical grid.
Operators
- setzaxis
-
Set z-axis
This operator sets the z-axis description of all variables with the same number of level as the new z-axis. - genlevelbounds
-
Generate level bounds
Generates the layer bounds of the z-axis.
Parameter
- zaxis
-
STRING Z-axis description file or name of the target z-axis
- zbot
-
FLOAT Specifying the bottom of the vertical column. Must have the same units as z-axis.
- ztop
-
FLOAT Specifying the top of the vertical column. Must have the same units as z-axis.
2.6.8 INVERT - Invert latitudes
Synopsis
invertlat infile outfile
Description
This operator inverts the latitudes of all fields on a rectilinear grid.
Example
To invert the latitudes of a 2D field from N->S to S->N use:
cdo invertlat infile outfile
2.6.9 INVERTLEV - Invert levels
Synopsis
invertlev infile outfile
Description
This operator inverts the levels of all 3D variables.
2.6.10 SHIFTXY - Shift field
Synopsis
<operator>,<nshift>,<cyclic>,<coord> infile outfile
Description
This module contains operators to shift all fields in x or y direction. All fields need to have the same horizontal rectilinear or curvilinear grid.
Operators
- shiftx
-
Shift x
Shifts all fields in x direction. - shifty
-
Shift y
Shifts all fields in y direction.
Parameter
- nshift
-
INTEGER Number of grid cells to shift (default: 1)
- cyclic
-
STRING If set, cells are filled up cyclic (default: missing value)
- coord
-
STRING If set, coordinates are also shifted
Example
To shift all input fields in the x direction by +1 cells and fill the new cells with missing value, use:
cdo shiftx infile outfile
To shift all input fields in the x direction by +1 cells and fill the new cells cyclic, use:
cdo shiftx,1,cyclic infile outfile
2.6.11 MASKREGION - Mask regions
Synopsis
maskregion,regions infile outfile
Description
Masks different regions of the input fields. The grid cells inside a region are untouched, the cells outside are set to missing value. Considered are only those grid cells with the grid center inside the regions. All input fields must have the same horizontal grid.
Regions can be defined by the user via an ASCII file. Each region consists of the geographic coordinates of a polygon. Each line of a polygon description file contains the longitude and latitude of one point. Each polygon description file can contain one or more polygons separated by a line with the character &.
Predefined regions of countries can be specified via the country codes. A country is specified with dcw:<CountryCode>. Country codes can be combined with the plus sign.
Parameter
- regions
-
STRING Comma-separated list of ASCII formatted files with different regions
Example
To mask the region with the longitudes from 120E to 90W and latitudes from 20N to 20S on all input fields use:
cdo maskregion,myregion infile outfile
For this example the description file of the region myregion should contain one polygon with the following four coordinates:
120 20 120 -20 270 -20 270 20
To mask the region of a country use the country code with data from the Digital Chart of the World. Here is an example for Spain with the country code ES:
cdo maskregion,dcw:ES infile outfile
2.6.12 MASKBOX - Mask a box
Synopsis
masklonlatbox,lon1,lon2,lat1,lat2 infile outfile
maskindexbox,idx1,idx2,idy1,idy2 infile outfile
Description
Masks grid cells inside a lon/lat or index box. The elements inside the box are untouched, the elements outside are set to missing value. All input fields need to have the same horizontal grid. Use sellonlatbox or selindexbox if only the data inside the box are needed.
Operators
- masklonlatbox
-
Mask a longitude/latitude box
Masks grid cells inside a lon/lat box. The user must specify the longitude and latitude of the edges of the box. Only those grid cells are considered whose grid center lies within the lon/lat box. For rotated lon/lat grids the parameters must be specified in rotated coordinates. - maskindexbox
-
Mask an index box
Masks grid cells within an index box. The user must specify the indices of the edges of the box. The index of the left edge can be greater then the one of the right edge. Use negative indexing to start from the end. The input grid must be a regular lon/lat or a 2D curvilinear grid.
Parameter
- lon1
-
FLOAT Western longitude
- lon2
-
FLOAT Eastern longitude
- lat1
-
FLOAT Southern or northern latitude
- lat2
-
FLOAT Northern or southern latitude
- idx1
-
INTEGER Index of first longitude
- idx2
-
INTEGER Index of last longitude
- idy1
-
INTEGER Index of first latitude
- idy2
-
INTEGER Index of last latitude
Example
To mask the region with the longitudes from 120E to 90W and latitudes from 20N to 20S on all input fields use:
cdo masklonlatbox,120,-90,20,-20 infile outfile
If the input dataset has fields on a regular Gaussian F16 grid, the same box can be masked with maskindexbox by:
cdo maskindexbox,23,48,13,20 infile outfile
2.6.13 SETBOX - Set a box to constant
Synopsis
setclonlatbox,c,lon1,lon2,lat1,lat2 infile outfile
setcindexbox,c,idx1,idx2,idy1,idy2 infile outfile
Description
Sets a box of the rectangularly understood field to a constant value. The elements outside the box are untouched, the elements inside are set to the given constant. All input fields need to have the same horizontal grid.
Operators
- setclonlatbox
-
Set a longitude/latitude box to constant
Sets the values of a longitude/latitude box to a constant value. The user has to give the longitudes and latitudes of the edges of the box. - setcindexbox
-
Set an index box to constant
Sets the values of an index box to a constant value. The user has to give the indices of the edges of the box. The index of the left edge can be greater than the one of the right edge.
Parameter
- c
-
FLOAT Constant
- lon1
-
FLOAT Western longitude
- lon2
-
FLOAT Eastern longitude
- lat1
-
FLOAT Southern or northern latitude
- lat2
-
FLOAT Northern or southern latitude
- idx1
-
INTEGER Index of first longitude
- idx2
-
INTEGER Index of last longitude
- idy1
-
INTEGER Index of first latitude
- idy2
-
INTEGER Index of last latitude
Example
To set all values in the region with the longitudes from 120E to 90W and latitudes from 20N to 20S to the constant value -1.23 use:
cdo setclonlatbox,-1.23,120,-90,20,-20 infile outfile
If the input dataset has fields on a regular Gaussian F16 grid, the same box can be set with setcindexbox by:
cdo setcindexbox,-1.23,23,48,13,20 infile outfile
2.6.14 ENLARGE - Enlarge fields
Synopsis
enlarge,grid infile outfile
Description
Enlarge all fields of infile to a user given horizontal grid. Normally only the last field element is used for the enlargement. If however the input and output grid are regular lon/lat grids, a zonal or meridional enlargement is possible. Zonal enlargement takes place, if the xsize of the input field is 1 and the ysize of both grids are the same. For meridional enlargement the ysize have to be 1 and the xsize of both grids should have the same size.
Parameter
- grid
-
STRING Target grid description file or name
Example
Assumed you want to add two datasets. The first dataset is a field on a global grid (n field elements) and the second dataset is a global mean (1 field element). Before you can add these two datasets the second dataset have to be enlarged to the grid size of the first dataset:
cdo enlarge,infile1 infile2 tmpfile cdo add infile1 tmpfile outfile
Or shorter using operator piping:
cdo add infile1 -enlarge,infile1 infile2 outfile
2.6.15 SETMISS - Set missing value
Synopsis
setmissval,newmiss infile outfile
setctomiss,c infile outfile
setmisstoc,c infile outfile
setrtomiss,rmin,rmax infile outfile
setvrange,rmin,rmax infile outfile
setmisstonn infile outfile
setmisstodis[,neighbors] infile outfile
Description
This module sets part of a field to missing value or missing values to a constant value. Which part of the field is set depends on the chosen operator.
Operators
- setmissval
-
Set a new missing value
o(t,x) =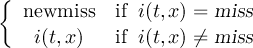
- setctomiss
-
Set constant to missing value
o(t,x) =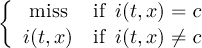
- setmisstoc
-
Set missing value to constant
o(t,x) =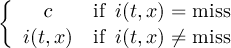
- setrtomiss
-
Set range to missing value
o(t,x) =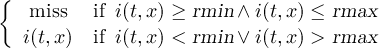
- setvrange
-
Set valid range
o(t,x) =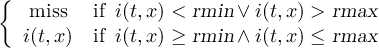
- setmisstonn
-
Set missing value to nearest neighbor
Set all missing values to the nearest non missing value.o(t,x) =
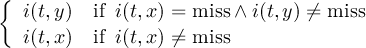
- setmisstodis
-
Set missing value to distance-weighted average
Set all missing values to the distance-weighted average of the nearest non missing values. The default number of nearest neighbors is 4.
Parameter
- neighbors
-
INTEGER Number of nearest neighbors
- newmiss
-
FLOAT New missing value
- c
-
FLOAT Constant
- rmin
-
FLOAT Lower bound
- rmax
-
FLOAT Upper bound
Example
setrtomiss
Assume an input dataset has one field with temperatures in the range from 246 to 304 Kelvin. To set all values below 273.15 Kelvin to missing value use:
cdo setrtomiss,0,273.15 infile outfile
Result of ’cdo info infile’:
-1 : Date Time Code Level Size Miss : Minimum Mean Maximum 1 : 1987-12-31 12:00:00 139 0 2048 0 : 246.27 276.75 303.71
Result of ’cdo info outfile’:
-1 : Date Time Code Level Size Miss : Minimum Mean Maximum 1 : 1987-12-31 12:00:00 139 0 2048 871 : 273.16 287.08 303.71
setmisstonn
Set all missing values to the nearest non missing value:
cdo setmisstonn infile outfile
Below is a schematic illustration of this example:
On the left side is input data with missing values in grey and on the right side the result with the filled missing values.
2.6.16 VERTFILLMISS - Vertical filling of missing values
Synopsis
vertfillmiss[,parameter] infile outfile
Description
This operator fills in vertical missing values. The method parameter can be used to select the filling method. The default method=nearest fills missing values with the nearest neighbor value. Other options are forward and backward to fill missing values by forward or backward propagation of values. Use the limit parameter to set the maximum number of consecutive missing values to fill and max_gaps to set the maximum number of gaps to fill.
Parameter
- method
-
STRING Fill method [nearest|linear|forward|backward] (default: nearest)
- limit
-
INTEGER The maximum number of consecutive missing values to fill (default: all)
- max_gaps
-
INTEGER The maximum number of gaps to fill (default: all)
2.6.17 TIMFILLMISS - Temporal filling of missing values
Synopsis
timfillmiss[,parameter] infile outfile
Description
This operator fills in temporally missing values. The method parameter can be used to select the filling method. The default method=nearest fills missing values with the nearest neighbor value. Other options are forward and backward to fill missing values by forward or backward propagation of values. Use the limit parameter to set the maximum number of consecutive missing values to fill and max_gaps to set the maximum number of gaps to fill.
Parameter
- method
-
STRING Fill method [nearest|linear|forward|backward] (default: nearest)
- limit
-
INTEGER The maximum number of consecutive missing values to fill (default: all)
- max_gaps
-
INTEGER The maximum number of gaps to fill (default: all)
2.6.18 SETGRIDCELL - Set the value of a grid cell
Synopsis
setgridcell,parameter infile outfile
Description
This operator sets the value of the selected grid cells. The grid cells can be selected by a comma-separated list of grid cell indices or a mask. The mask is read from a data file, which may contain only one field. If no grid cells are selected, all values are set.
Parameter
- value
-
FLOAT Value of the grid cell
- cell
-
INTEGER Comma-separated list of grid cell indices
- mask
-
STRING Name of the data file which contains the mask
2.7 Arithmetic
This section contains modules to arithmetically process datasets.
Here is a short overview of all operators in this section:
| expr | Evaluate expressions |
| exprf | Evaluate expressions script |
| aexpr | Evaluate expressions and append results |
| aexprf | Evaluate expression script and append results |
| abs | Absolute value |
| int | Integer value |
| nint | Nearest integer value |
| pow | Power |
| sqr | Square |
| sqrt | Square root |
| exp | Exponential |
| ln | Natural logarithm |
| log10 | Base 10 logarithm |
| sin | Sine |
| cos | Cosine |
| tan | Tangent |
| asin | Arc sine |
| acos | Arc cosine |
| atan | Arc tangent |
| reci | Reciprocal value |
| not | Logical NOT |
| addc | Add a constant |
| subc | Subtract a constant |
| mulc | Multiply with a constant |
| divc | Divide by a constant |
| minc | Minimum of a field and a constant |
| maxc | Maximum of a field and a constant |
| add | Add two fields |
| sub | Subtract two fields |
| mul | Multiply two fields |
| div | Divide two fields |
| min | Minimum of two fields |
| max | Maximum of two fields |
| atan2 | Arc tangent of two fields |
| setmiss | Set missing values |
| dayadd | Add daily time series |
| daysub | Subtract daily time series |
| daymul | Multiply daily time series |
| daydiv | Divide daily time series |
| monadd | Add monthly time series |
| monsub | Subtract monthly time series |
| monmul | Multiply monthly time series |
| mondiv | Divide monthly time series |
| yearadd | Add yearly time series |
| yearsub | Subtract yearly time series |
| yearmul | Multiply yearly time series |
| yeardiv | Divide yearly time series |
| yhouradd | Add multi-year hourly time series |
| yhoursub | Subtract multi-year hourly time series |
| yhourmul | Multiply multi-year hourly time series |
| yhourdiv | Divide multi-year hourly time series |
| ydayadd | Add multi-year daily time series |
| ydaysub | Subtract multi-year daily time series |
| ydaymul | Multiply multi-year daily time series |
| ydaydiv | Divide multi-year daily time series |
| ymonadd | Add multi-year monthly time series |
| ymonsub | Subtract multi-year monthly time series |
| ymonmul | Multiply multi-year monthly time series |
| ymondiv | Divide multi-year monthly time series |
| yseasadd | Add multi-year seasonal time series |
| yseassub | Subtract multi-year seasonal time series |
| yseasmul | Multiply multi-year seasonal time series |
| yseasdiv | Divide multi-year seasonal time series |
| muldpm | Multiply with days per month |
| divdpm | Divide by days per month |
| muldpy | Multiply with days per year |
| divdpy | Divide by days per year |
| mulcoslat | Multiply with the cosine of the latitude |
| divcoslat | Divide by cosine of the latitude |
2.7.1 EXPR - Evaluate expressions
Synopsis
expr,instr infile outfile
exprf,filename infile outfile
aexpr,instr infile outfile
aexprf,filename infile outfile
Description
This module arithmetically processes every timestep of the input dataset. Each individual assignment statement have to end with a semi-colon. The special key _ALL_ is used as a template. A statement with a template is replaced for all variable names. Unlike regular variables, temporary variables are never written to the output stream. To define a temporary variable simply prefix the variable name with an underscore (e.g. _varname) when the variable is declared.
The following operators are supported:
| Operator | Meaning | Example | Result |
| = | assignment | x = y | Assigns y to x |
| + | addition | x + y | Sum of x and y |
| - | subtraction | x - y | Difference of x and y |
| * | multiplication | x * y | Product of x and y |
| / | division | x / y | Quotient of x and y |
 | exponentiation | x  y y | Exponentiates x with y |
| == | equal to | x == y | 1, if x equal to y; else 0 |
| != | not equal to | x != y | 1, if x not equal to y; else 0 |
| > | greater than | x > y | 1, if x greater than y; else 0 |
| < | less than | x < y | 1, if x less than y; else 0 |
| >= | greater equal | x >= y | 1, if x greater equal y; else 0 |
| <= | less equal | x <= y | 1, if x less equal y; else 0 |
| <=> | less equal greater | x <=> y | -1, if x less y; 1, if x greater y; else 0 |
| && | logical AND | x && y | 1, if x and y not equal 0; else 0 |
| || | logical OR | x || y | 1, if x or y not equal 0; else 0 |
| ! | logical NOT | !x | 1, if x equal 0; else 0 |
| ?: | ternary conditional | x ? y : z | y, if x not equal 0, else z |
The following functions are supported:
Math intrinsics:
- abs(x)
-
Absolute value of x
- floor(x)
-
Round to largest integral value not greater than x
- ceil(x)
-
Round to smallest integral value not less than x
- float(x)
-
32-bit float value of x
- int(x)
-
Integer value of x
- nint(x)
-
Nearest integer value of x
- sqr(x)
-
Square of x
- sqrt(x)
-
Square Root of x
- exp(x)
-
Exponential of x
- ln(x)
-
Natural logarithm of x
- log10(x)
-
Base 10 logarithm of x
- sin(x)
-
Sine of x, where x is specified in radians
- cos(x)
-
Cosine of x, where x is specified in radians
- tan(x)
-
Tangent of x, where x is specified in radians
- asin(x)
-
Arc-sine of x, where x is specified in radians
- acos(x)
-
Arc-cosine of x, where x is specified in radians
- atan(x)
-
Arc-tangent of x, where x is specified in radians
- sinh(x)
-
Hyperbolic sine of x, where x is specified in radians
- cosh(x)
-
Hyperbolic cosine of x, where x is specified in radians
- tanh(x)
-
Hyperbolic tangent of x, where x is specified in radians
- asinh(x)
-
Inverse hyperbolic sine of x, where x is specified in radians
- acosh(x)
-
Inverse hyperbolic cosine of x, where x is specified in radians
- atanh(x)
-
Inverse hyperbolic tangent of x, where x is specified in radians
- rad(x)
-
Convert x from degrees to radians
- deg(x)
-
Convert x from radians to degrees
- rand(x)
-
Replace x by pseudo-random numbers in the range of 0 to 1
- isMissval(x)
-
Returns 1 where x is missing
- mod(x,y)
-
Floating-point remainder of x/ y
- min(x,y)
-
Minimum value of x and y
- max(x,y)
-
Maximum value of x and y
- pow(x,y)
-
Power function
- hypot(x,y)
-
Euclidean distance function, sqrt(x*x + y*y)
- atan2(x,y)
-
Arc tangent function of y/x, using signs to determine quadrants
- trimrel(x,kb)
-
Trim relative precision to kb keep-bits. Max relative error to a value | result - x | / x < 2**(-1-kb) for any finite x. trimrel is a non-decreasing function of x. Loosely follows A5 of https://gmd.copernicus.org/articles/14/377/2021/
- trimabs(x,err)
-
Trim absolute precision introducing given max. absolute error | result - x | < err, trimabs is a non-decreasing function of x. Loosely follows A6 of https://gmd.copernicus.org/articles/14/377/2021/
Coordinates:
- clon(x)
-
Longitude coordinate of x (available only if x has geographical coordinates)
- clat(x)
-
Latitude coordinate of x (available only if x has geographical coordinates)
- gridarea(x)
-
Grid cell area of x (available only if x has geographical coordinates)
- gridindex(x)
-
Grid cell indices of x
- clev(x)
-
Level coordinate of x (0, if x is a 2D surface variable)
- clevidx(x)
-
Level index of x (0, if x is a 2D surface variable)
- cthickness(x)
-
Layer thickness, upper minus lower level bound of x (1, if level bounds are missing)
- ctimestep()
-
Timestep number (1 to N)
- cdate()
-
Verification date as YYYYMMDD
- ctime()
-
Verification time as HHMMSS.millisecond
- cdeltat()
-
Difference between current and last timestep in seconds
- cday()
-
Day as DD
- cmonth()
-
Month as MM
- cyear()
-
Year as YYYY
- csecond()
-
Second as SS.millisecond
- cminute()
-
Minute as MM
- chour()
-
Hour as HH
Constants:
- ngp(x)
-
Number of horizontal grid points
- nlev(x)
-
Number of vertical levels
- size(x)
-
Total number of elements (ngp(x)*nlev(x))
- missval(x)
-
Returns the missing value of variable x
Statistics over a field:
fldmin(x), fldmax(x), fldrange(x), fldsum(x), fldmean(x), fldavg(x), fldstd(x), fldstd1(x), fldvar(x), fldvar1(x), fldskew(x), fldkurt(x), fldmedian(x)
Zonal statistics for regular 2D grids:
zonmin(x), zonmax(x), zonrange(x), zonsum(x), zonmean(x), zonavg(x), zonstd(x), zonstd1(x), zonvar(x), zonvar1(x), zonskew(x), zonkurt(x), zonmedian(x)
Vertical statistics:
vertmin(x), vertmax(x), vertrange(x), vertsum(x), vertmean(x), vertavg(x), vertstd(x), vertstd1(x), vertvar(x), vertvar1(x)
Miscellaneous:
- sellevel(x,k)
-
Select level k of variable x
- sellevidx(x,k)
-
Select level index k of variable x
- sellevelrange(x,k1,k2)
-
Select all levels of variable x in the range k1 to k2
- sellevidxrange(x,k1,k2)
-
Select all level indices of variable x in the range k1 to k2
- remove(x)
-
Remove variable x from output stream
Operators
- expr
-
Evaluate expressions
The processing instructions are read from the parameter. - exprf
-
Evaluate expressions script
Contrary to expr the processing instructions are read from a file. - aexpr
-
Evaluate expressions and append results
Same as expr, but keep input variables and append results - aexprf
-
Evaluate expression script and append results
Same as exprf, but keep input variables and append results
Parameter
- instr
-
STRING Processing instructions (need to be ’quoted’ in most cases)
- filename
-
STRING File with processing instructions
Note
If the input stream contains duplicate entries of the same variable name then the last one is used.
Example
Assume an input dataset contains at least the variables ’aprl’, ’aprc’ and ’ts’. To create a new variable ’var1’ with the sum of ’aprl’ and ’aprc’ and a variable ’var2’ which convert the temperature ’ts’ from Kelvin to Celsius use:
cdo expr,’var1=aprl+aprc;var2=ts-273.15;’ infile outfile
The same example, but the instructions are read from a file:
cdo exprf,myexpr infile outfile
The file myexpr contains:
var1 = aprl + aprc; var2 = ts - 273.15;
2.7.2 MATH - Mathematical functions
Synopsis
<operator> infile outfile
Description
This module contains some standard mathematical functions. All trigonometric functions calculate with radians.
Operators
- abs
-
Absolute value
o(t,x) = abs(i(t,x)) - int
-
Integer value
o(t,x) = int(i(t,x)) - nint
-
Nearest integer value
o(t,x) = nint(i(t,x)) - pow
-
Power
o(t,x) = i(t,x)y - sqr
-
Square
o(t,x) = i(t,x)2 - sqrt
-
Square root
o(t,x) =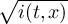
- exp
-
Exponential
o(t,x) = ei(t,x) - ln
-
Natural logarithm
o(t,x) = ln(i(t,x)) - log10
-
Base 10 logarithm
o(t,x) = log 10(i(t,x)) - sin
-
Sine
o(t,x) = sin(i(t,x)) - cos
-
Cosine
o(t,x) = cos(i(t,x)) - tan
-
Tangent
o(t,x) = tan(i(t,x)) - asin
-
Arc sine
o(t,x) = arcsin(i(t,x)) - acos
-
Arc cosine
o(t,x) = arccos(i(t,x)) - atan
-
Arc tangent
o(t,x) = arctan(i(t,x)) - reci
-
Reciprocal value
o(t,x) = 1∕i(t,x) - not
-
Logical NOT
o(t,x) = 1,ifxequal0;else0
Example
To calculate the square root for all field elements use:
cdo sqrt infile outfile
2.7.3 ARITHC - Arithmetic with a constant
Synopsis
<operator>,c infile outfile
Description
This module performs simple arithmetic with all field elements of a dataset and a constant. The fields in outfile inherit the meta data from infile.
Operators
- addc
-
Add a constant
o(t,x) = i(t,x) + c - subc
-
Subtract a constant
o(t,x) = i(t,x) - c - mulc
-
Multiply with a constant
o(t,x) = i(t,x) * c - divc
-
Divide by a constant
o(t,x) = i(t,x)∕c - minc
-
Minimum of a field and a constant
o(t,x) = min(i(t,x),c) - maxc
-
Maximum of a field and a constant
o(t,x) = max(i(t,x),c)
Parameter
- c
-
FLOAT Constant
Example
To sum all input fields with the constant -273.15 use:
cdo addc,-273.15 infile outfile
2.7.4 ARITH - Arithmetic on two datasets
Synopsis
<operator> infile1 infile2 outfile
Description
This module performs simple arithmetic of two datasets. The number of fields in infile1 should be the same as in infile2. The fields in outfile inherit the meta data from infile1. All operators in this module simply process one field after the other from the two input files. Neither the order of the variables nor the date is checked. One of the input files can contain only one timestep or one variable.
Operators
- add
-
Add two fields
o(t,x) = i1(t,x) + i2(t,x) - sub
-
Subtract two fields
o(t,x) = i1(t,x) - i2(t,x) - mul
-
Multiply two fields
o(t,x) = i1(t,x) * i2(t,x) - div
-
Divide two fields
o(t,x) = i1(t,x)∕i2(t,x) - min
-
Minimum of two fields
o(t,x) = min(i1(t,x),i2(t,x)) - max
-
Maximum of two fields
o(t,x) = max(i1(t,x),i2(t,x)) - atan2
-
Arc tangent of two fields
The atan2 operator calculates the arc tangent of two fields. The result is in radians, which is between -PI and PI (inclusive).o(t,x) = atan2(i1(t,x),i2(t,x))
- setmiss
-
Set missing values
Sets missing values of infile1 to values from infile2.
Example
To sum all fields of the first input file with the corresponding fields of the second input file use:
cdo add infile1 infile2 outfile
2.7.5 DAYARITH - Daily arithmetic
Synopsis
<operator> infile1 infile2 outfile
Description
This module performs simple arithmetic of a time series and one timestep with the same day, month and year. For each field in infile1 the corresponding field of the timestep in infile2 with the same day, month and year is used. The input files need to have the same structure with the same variables. Usually infile2 is generated by an operator of the module DAYSTAT.
Operators
- dayadd
-
Add daily time series
Adds a time series and a daily time series. - daysub
-
Subtract daily time series
Subtracts a time series and a daily time series. - daymul
-
Multiply daily time series
Multiplies a time series and a daily time series. - daydiv
-
Divide daily time series
Divides a time series and a daily time series.
Example
To subtract a daily time average from a time series use:
cdo daysub infile -dayavg infile outfile
2.7.6 MONARITH - Monthly arithmetic
Synopsis
<operator> infile1 infile2 outfile
Description
This module performs simple arithmetic of a time series and one timestep with the same month and year. For each field in infile1 the corresponding field of the timestep in infile2 with the same month and year is used. The input files need to have the same structure with the same variables. Usually infile2 is generated by an operator of the module MONSTAT.
Operators
- monadd
-
Add monthly time series
Adds a time series and a monthly time series. - monsub
-
Subtract monthly time series
Subtracts a time series and a monthly time series. - monmul
-
Multiply monthly time series
Multiplies a time series and a monthly time series. - mondiv
-
Divide monthly time series
Divides a time series and a monthly time series.
Example
To subtract a monthly time average from a time series use:
cdo monsub infile -monavg infile outfile
2.7.7 YEARARITH - Yearly arithmetic
Synopsis
<operator> infile1 infile2 outfile
Description
This module performs simple arithmetic of a time series and one timestep with the same year. For each field in infile1 the corresponding field of the timestep in infile2 with the same year is used. The header information in infile1 have to be the same as in infile2. Usually infile2 is generated by an operator of the module YEARSTAT.
Operators
- yearadd
-
Add yearly time series
Adds a time series and a yearly time series. - yearsub
-
Subtract yearly time series
Subtracts a time series and a yearly time series. - yearmul
-
Multiply yearly time series
Multiplies a time series and a yearly time series. - yeardiv
-
Divide yearly time series
Divides a time series and a yearly time series.
Example
To subtract a yearly time average from a time series use:
cdo yearsub infile -yearavg infile outfile
2.7.8 YHOURARITH - Multi-year hourly arithmetic
Synopsis
<operator> infile1 infile2 outfile
Description
This module performs simple arithmetic of a time series and one timestep with the same hour and day of year. For each field in infile1 the corresponding field of the timestep in infile2 with the same hour and day of year is used. The input files need to have the same structure with the same variables. Usually infile2 is generated by an operator of the module YHOURSTAT.
Operators
- yhouradd
-
Add multi-year hourly time series
Adds a time series and a multi-year hourly time series. - yhoursub
-
Subtract multi-year hourly time series
Subtracts a time series and a multi-year hourly time series. - yhourmul
-
Multiply multi-year hourly time series
Multiplies a time series and a multi-year hourly time series. - yhourdiv
-
Divide multi-year hourly time series
Divides a time series and a multi-year hourly time series.
Example
To subtract a multi-year hourly time average from a time series use:
cdo yhoursub infile -yhouravg infile outfile
2.7.9 YDAYARITH - Multi-year daily arithmetic
Synopsis
<operator> infile1 infile2 outfile
Description
This module performs simple arithmetic of a time series and one timestep with the same day of year. For each field in infile1 the corresponding field of the timestep in infile2 with the same day of year is used. The input files need to have the same structure with the same variables. Usually infile2 is generated by an operator of the module YDAYSTAT.
Operators
- ydayadd
-
Add multi-year daily time series
Adds a time series and a multi-year daily time series. - ydaysub
-
Subtract multi-year daily time series
Subtracts a time series and a multi-year daily time series. - ydaymul
-
Multiply multi-year daily time series
Multiplies a time series and a multi-year daily time series. - ydaydiv
-
Divide multi-year daily time series
Divides a time series and a multi-year daily time series.
Example
To subtract a multi-year daily time average from a time series use:
cdo ydaysub infile -ydayavg infile outfile
2.7.10 YMONARITH - Multi-year monthly arithmetic
Synopsis
<operator> infile1 infile2 outfile
Description
This module performs simple arithmetic of a time series and one timestep with the same month of year. For each field in infile1 the corresponding field of the timestep in infile2 with the same month of year is used. The input files need to have the same structure with the same variables. Usually infile2 is generated by an operator of the module YMONSTAT.
Operators
- ymonadd
-
Add multi-year monthly time series
Adds a time series and a multi-year monthly time series. - ymonsub
-
Subtract multi-year monthly time series
Subtracts a time series and a multi-year monthly time series. - ymonmul
-
Multiply multi-year monthly time series
Multiplies a time series with a multi-year monthly time series. - ymondiv
-
Divide multi-year monthly time series
Divides a time series by a multi-year monthly time series.
Example
To subtract a multi-year monthly time average from a time series use:
cdo ymonsub infile -ymonavg infile outfile
2.7.11 YSEASARITH - Multi-year seasonal arithmetic
Synopsis
<operator> infile1 infile2 outfile
Description
This module performs simple arithmetic of a time series and one timestep with the same season. For each field in infile1 the corresponding field of the timestep in infile2 with the same season is used. The input files need to have the same structure with the same variables. Usually infile2 is generated by an operator of the module YSEASSTAT.
Operators
- yseasadd
-
Add multi-year seasonal time series
Adds a time series and a multi-year seasonal time series. - yseassub
-
Subtract multi-year seasonal time series
Subtracts a time series and a multi-year seasonal time series. - yseasmul
-
Multiply multi-year seasonal time series
Multiplies a time series and a multi-year seasonal time series. - yseasdiv
-
Divide multi-year seasonal time series
Divides a time series and a multi-year seasonal time series.
Example
To subtract a multi-year seasonal time average from a time series use:
cdo yseassub infile -yseasavg infile outfile
2.7.12 ARITHDAYS - Arithmetic with days
Synopsis
<operator> infile outfile
Description
This module multiplies or divides each timestep of a dataset with the corresponding days per month or days per year. The result of these functions depends on the used calendar of the input data.
Operators
- muldpm
-
Multiply with days per month
o(t,x) = i(t,x) * days_per_month - divdpm
-
Divide by days per month
o(t,x) = i(t,x)∕days_per_month - muldpy
-
Multiply with days per year
o(t,x) = i(t,x) * days_per_year - divdpy
-
Divide by days per year
o(t,x) = i(t,x)∕days_per_year
2.7.13 ARITHLAT - Arithmetic with latitude
Synopsis
<operator> infile outfile
Description
This module multiplies or divides each field element with the cosine of the latitude.
Operators
- mulcoslat
-
Multiply with the cosine of the latitude
o(t,x) = i(t,x) * cos(latitude(x)) - divcoslat
-
Divide by cosine of the latitude
o(t,x) = i(t,x)∕cos(latitude(x))
2.8 Statistical values
This section contains modules to compute statistical values of datasets. In this program there is the different
notion of "mean" and "average" to distinguish two different kinds of treatment of missing values. While computing
the mean, only the not missing values are considered to belong to the sample with the side effect of a probably
reduced sample size. Computing the average is just adding the sample members and divide the result by the
sample size. For example, the mean of 1, 2, miss and 3 is (1+2+3)/3 = 2, whereas the average is
(1+2+miss+3)/4 = miss/4 = miss. If there are no missing values in the sample, the average and the mean are
identical.
CDO is using the verification time to identify the time range for temporal statistics. The time bounds are never
used!
In this section the abbreviations as in the following table are used:
![Skewness ∑n --
skew --i=1(xi --x)∕n
s3
Kurtosis ∑n (x - x)4∕n
kurt --i=1--i4-------
s
Cumulative Ranked ∫ ∞
Probability Score [H (x1)- cdf({x2...xn})|r]2dr
crps -∞](cdo41x.png)
 being the cumulative distribution function of
being the cumulative distribution function of  at
at

and
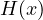 the Heavyside function jumping at
the Heavyside function jumping at  .
. | timcumsum | Cumulative sum over all timesteps |
| consecsum | Consecutive Sum |
| consects | Consecutive Timesteps |
| varsmin | Variables minimum |
| varsmax | Variables maximum |
| varsrange | Variables range |
| varssum | Variables sum |
| varsmean | Variables mean |
| varsavg | Variables average |
| varsstd | Variables standard deviation |
| varsstd1 | Variables standard deviation (n-1) |
| varsvar | Variables variance |
| varsvar1 | Variables variance (n-1) |
| ensmin | Ensemble minimum |
| ensmax | Ensemble maximum |
| ensrange | Ensemble range |
| enssum | Ensemble sum |
| ensmean | Ensemble mean |
| ensavg | Ensemble average |
| ensstd | Ensemble standard deviation |
| ensstd1 | Ensemble standard deviation (n-1) |
| ensvar | Ensemble variance |
| ensvar1 | Ensemble variance (n-1) |
| ensskew | Ensemble skewness |
| enskurt | Ensemble kurtosis |
| ensmedian | Ensemble median |
| enspctl | Ensemble percentiles |
| ensrkhistspace | Ranked Histogram averaged over space |
| ensrkhisttime | Ranked Histogram averaged over time |
| ensroc | Ensemble Receiver Operating characteristics |
| enscrps | Ensemble CRPS and decomposition |
| ensbrs | Ensemble Brier score |
| fldmin | Field minimum |
| fldmax | Field maximum |
| fldrange | Field range |
| fldsum | Field sum |
| fldint | Field integral |
| fldmean | Field mean |
| fldavg | Field average |
| fldstd | Field standard deviation |
| fldstd1 | Field standard deviation (n-1) |
| fldvar | Field variance |
| fldvar1 | Field variance (n-1) |
| fldskew | Field skewness |
| fldkurt | Field kurtosis |
| fldmedian | Field median |
| fldcount | Field count |
| fldpctl | Field percentiles |
| zonmin | Zonal minimum |
| zonmax | Zonal maximum |
| zonrange | Zonal range |
| zonsum | Zonal sum |
| zonmean | Zonal mean |
| zonavg | Zonal average |
| zonstd | Zonal standard deviation |
| zonstd1 | Zonal standard deviation (n-1) |
| zonvar | Zonal variance |
| zonvar1 | Zonal variance (n-1) |
| zonskew | Zonal skewness |
| zonkurt | Zonal kurtosis |
| zonmedian | Zonal median |
| zonpctl | Zonal percentiles |
| mermin | Meridional minimum |
| mermax | Meridional maximum |
| merrange | Meridional range |
| mersum | Meridional sum |
| mermean | Meridional mean |
| meravg | Meridional average |
| merstd | Meridional standard deviation |
| merstd1 | Meridional standard deviation (n-1) |
| mervar | Meridional variance |
| mervar1 | Meridional variance (n-1) |
| merskew | Meridional skewness |
| merkurt | Meridional kurtosis |
| mermedian | Meridional median |
| merpctl | Meridional percentiles |
| gridboxmin | Gridbox minimum |
| gridboxmax | Gridbox maximum |
| gridboxrange | Gridbox range |
| gridboxsum | Gridbox sum |
| gridboxmean | Gridbox mean |
| gridboxavg | Gridbox average |
| gridboxstd | Gridbox standard deviation |
| gridboxstd1 | Gridbox standard deviation (n-1) |
| gridboxvar | Gridbox variance |
| gridboxvar1 | Gridbox variance (n-1) |
| gridboxskew | Gridbox skewness |
| gridboxkurt | Gridbox kurtosis |
| gridboxmedian | Gridbox median |
| remapmin | Remap minimum |
| remapmax | Remap maximum |
| remaprange | Remap range |
| remapsum | Remap sum |
| remapmean | Remap mean |
| remapavg | Remap average |
| remapstd | Remap standard deviation |
| remapstd1 | Remap standard deviation (n-1) |
| remapvar | Remap variance |
| remapvar1 | Remap variance (n-1) |
| remapskew | Remap skewness |
| remapkurt | Remap kurtosis |
| remapmedian | Remap median |
| vertmin | Vertical minimum |
| vertmax | Vertical maximum |
| vertrange | Vertical range |
| vertsum | Vertical sum |
| vertmean | Vertical mean |
| vertavg | Vertical average |
| vertstd | Vertical standard deviation |
| vertstd1 | Vertical standard deviation (n-1) |
| vertvar | Vertical variance |
| vertvar1 | Vertical variance (n-1) |
| timselmin | Time selection minimum |
| timselmax | Time selection maximum |
| timselrange | Time selection range |
| timselsum | Time selection sum |
| timselmean | Time selection mean |
| timselavg | Time selection average |
| timselstd | Time selection standard deviation |
| timselstd1 | Time selection standard deviation (n-1) |
| timselvar | Time selection variance |
| timselvar1 | Time selection variance (n-1) |
| timselpctl | Time range percentiles |
| runmin | Running minimum |
| runmax | Running maximum |
| runrange | Running range |
| runsum | Running sum |
| runmean | Running mean |
| runavg | Running average |
| runstd | Running standard deviation |
| runstd1 | Running standard deviation (n-1) |
| runvar | Running variance |
| runvar1 | Running variance (n-1) |
| runpctl | Running percentiles |
| timmin | Time minimum |
| timmax | Time maximum |
| timminidx | Index of time minimum |
| timmaxidx | Index of time maximum |
| timrange | Time range |
| timsum | Time sum |
| timmean | Time mean |
| timavg | Time average |
| timstd | Time standard deviation |
| timstd1 | Time standard deviation (n-1) |
| timvar | Time variance |
| timvar1 | Time variance (n-1) |
| timpctl | Time percentiles |
| hourmin | Hourly minimum |
| hourmax | Hourly maximum |
| hourrange | Hourly range |
| hoursum | Hourly sum |
| hourmean | Hourly mean |
| houravg | Hourly average |
| hourstd | Hourly standard deviation |
| hourstd1 | Hourly standard deviation (n-1) |
| hourvar | Hourly variance |
| hourvar1 | Hourly variance (n-1) |
| hourpctl | Hourly percentiles |
| daymin | Daily minimum |
| daymax | Daily maximum |
| dayrange | Daily range |
| daysum | Daily sum |
| daymean | Daily mean |
| dayavg | Daily average |
| daystd | Daily standard deviation |
| daystd1 | Daily standard deviation (n-1) |
| dayvar | Daily variance |
| dayvar1 | Daily variance (n-1) |
| daypctl | Daily percentiles |
| monmin | Monthly minimum |
| monmax | Monthly maximum |
| monrange | Monthly range |
| monsum | Monthly sum |
| monmean | Monthly mean |
| monavg | Monthly average |
| monstd | Monthly standard deviation |
| monstd1 | Monthly standard deviation (n-1) |
| monvar | Monthly variance |
| monvar1 | Monthly variance (n-1) |
| monpctl | Monthly percentiles |
| yearmonmean | Yearly mean from monthly data |
| yearmin | Yearly minimum |
| yearmax | Yearly maximum |
| yearminidx | Index of yearly minimum |
| yearmaxidx | Index of yearly maximum |
| yearrange | Yearly range |
| yearsum | Yearly sum |
| yearmean | Yearly mean |
| yearavg | Yearly average |
| yearstd | Yearly standard deviation |
| yearstd1 | Yearly standard deviation (n-1) |
| yearvar | Yearly variance |
| yearvar1 | Yearly variance (n-1) |
| yearpctl | Yearly percentiles |
| seasmin | Seasonal minimum |
| seasmax | Seasonal maximum |
| seasrange | Seasonal range |
| seassum | Seasonal sum |
| seasmean | Seasonal mean |
| seasavg | Seasonal average |
| seasstd | Seasonal standard deviation |
| seasstd1 | Seasonal standard deviation (n-1) |
| seasvar | Seasonal variance |
| seasvar1 | Seasonal variance (n-1) |
| seaspctl | Seasonal percentiles |
| yhourmin | Multi-year hourly minimum |
| yhourmax | Multi-year hourly maximum |
| yhourrange | Multi-year hourly range |
| yhoursum | Multi-year hourly sum |
| yhourmean | Multi-year hourly mean |
| yhouravg | Multi-year hourly average |
| yhourstd | Multi-year hourly standard deviation |
| yhourstd1 | Multi-year hourly standard deviation (n-1) |
| yhourvar | Multi-year hourly variance |
| yhourvar1 | Multi-year hourly variance (n-1) |
| dhourmin | Multi-day hourly minimum |
| dhourmax | Multi-day hourly maximum |
| dhourrange | Multi-day hourly range |
| dhoursum | Multi-day hourly sum |
| dhourmean | Multi-day hourly mean |
| dhouravg | Multi-day hourly average |
| dhourstd | Multi-day hourly standard deviation |
| dhourstd1 | Multi-day hourly standard deviation (n-1) |
| dhourvar | Multi-day hourly variance |
| dhourvar1 | Multi-day hourly variance (n-1) |
| dminutemin | Multi-day by the minute minimum |
| dminutemax | Multi-day by the minute maximum |
| dminuterange | Multi-day by the minute range |
| dminutesum | Multi-day by the minute sum |
| dminutemean | Multi-day by the minute mean |
| dminuteavg | Multi-day by the minute average |
| dminutestd | Multi-day by the minute standard deviation |
| dminutestd1 | Multi-day by the minute standard deviation (n-1) |
| dminutevar | Multi-day by the minute variance |
| dminutevar1 | Multi-day by the minute variance (n-1) |
| ydaymin | Multi-year daily minimum |
| ydaymax | Multi-year daily maximum |
| ydayrange | Multi-year daily range |
| ydaysum | Multi-year daily sum |
| ydaymean | Multi-year daily mean |
| ydayavg | Multi-year daily average |
| ydaystd | Multi-year daily standard deviation |
| ydaystd1 | Multi-year daily standard deviation (n-1) |
| ydayvar | Multi-year daily variance |
| ydayvar1 | Multi-year daily variance (n-1) |
| ydaypctl | Multi-year daily percentiles |
| ymonmin | Multi-year monthly minimum |
| ymonmax | Multi-year monthly maximum |
| ymonrange | Multi-year monthly range |
| ymonsum | Multi-year monthly sum |
| ymonmean | Multi-year monthly mean |
| ymonavg | Multi-year monthly average |
| ymonstd | Multi-year monthly standard deviation |
| ymonstd1 | Multi-year monthly standard deviation (n-1) |
| ymonvar | Multi-year monthly variance |
| ymonvar1 | Multi-year monthly variance (n-1) |
| ymonpctl | Multi-year monthly percentiles |
| yseasmin | Multi-year seasonal minimum |
| yseasmax | Multi-year seasonal maximum |
| yseasrange | Multi-year seasonal range |
| yseassum | Multi-year seasonal sum |
| yseasmean | Multi-year seasonal mean |
| yseasavg | Multi-year seasonal average |
| yseasstd | Multi-year seasonal standard deviation |
| yseasstd1 | Multi-year seasonal standard deviation (n-1) |
| yseasvar | Multi-year seasonal variance |
| yseasvar1 | Multi-year seasonal variance (n-1) |
| yseaspctl | Multi-year seasonal percentiles |
| ydrunmin | Multi-year daily running minimum |
| ydrunmax | Multi-year daily running maximum |
| ydrunsum | Multi-year daily running sum |
| ydrunmean | Multi-year daily running mean |
| ydrunavg | Multi-year daily running average |
| ydrunstd | Multi-year daily running standard deviation |
| ydrunstd1 | Multi-year daily running standard deviation (n-1) |
| ydrunvar | Multi-year daily running variance |
| ydrunvar1 | Multi-year daily running variance (n-1) |
| ydrunpctl | Multi-year daily running percentiles |
2.8.1 TIMCUMSUM - Cumulative sum over all timesteps
Synopsis
timcumsum infile outfile
Description
The timcumsum operator calculates the cumulative sum over all timesteps. Missing values are treated as numeric zero when summing.
o(t,x) = sum{i(t′,x),0 ≤ t′≤ t}
2.8.2 CONSECSTAT - Consecute timestep periods
Synopsis
<operator> infile outfile
Description
This module computes periods over all timesteps in infile where a certain property is valid. The property can be chosen by creating a mask from the original data, which is the expected input format for operators of this module. Depending on the operator full information about each period or just its length and ending date are computed.
Operators
- consecsum
-
Consecutive Sum
This operator computes periods of consecutive timesteps similar to a runsum, but periods are finished, when the mask value is 0. That way multiple periods can be found. Timesteps from the input are preserved. Missing values are handled like 0, i.e. finish periods of consecutive timesteps. - consects
-
Consecutive Timesteps
In contrast to the operator above consects only computes the length of each period together with its last timestep. To be able to perform statistical analysis like min, max or mean, everything else is set to missing value.
Example
For a given time series of daily temperatures, the periods of summer days can be calculated with inplace maskting the input field:
cdo consects -gtc,20.0 infile1 outfile
2.8.3 VARSSTAT - Statistical values over all variables
Synopsis
<operator> infile outfile
Description
This module computes statistical values over all variables for each timestep. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation is written to outfile. All input variables need to have the same gridsize and the same number of levels.
Operators
- varsmin
-
Variables minimum
For every timestep the minimum over all variables is computed. - varsmax
-
Variables maximum
For every timestep the maximum over all variables is computed. - varsrange
-
Variables range
For every timestep the range over all variables is computed. - varssum
-
Variables sum
For every timestep the sum over all variables is computed. - varsmean
-
Variables mean
For every timestep the mean over all variables is computed. - varsavg
-
Variables average
For every timestep the average over all variables is computed. - varsstd
-
Variables standard deviation
For every timestep the standard deviation over all variables is computed. Normalize by n. - varsstd1
-
Variables standard deviation (n-1)
For every timestep the standard deviation over all variables is computed. Normalize by (n-1). - varsvar
-
Variables variance
For every timestep the variance over all variables is computed. Normalize by n. - varsvar1
-
Variables variance (n-1)
For every timestep the variance over all variables is computed. Normalize by (n-1).
2.8.4 ENSSTAT - Statistical values over an ensemble
Synopsis
<operator> infiles outfile
enspctl,p infiles outfile
Description
This module computes statistical values over an ensemble of input files. Depending on the chosen operator, the minimum, maximum, range, sum, average, standard deviation, variance, skewness, kurtosis, median or a certain percentile over all input files is written to outfile. All input files need to have the same structure with the same variables. The date information of a timestep in outfile is the date of the first input file.
Operators
- ensmin
-
Ensemble minimum
o(t,x) = min{i1(t,x),i2(t,x), ,in(t,x)}
,in(t,x)}
- ensmax
-
Ensemble maximum
o(t,x) = max{i1(t,x),i2(t,x), ,in(t,x)}
,in(t,x)}
- ensrange
-
Ensemble range
o(t,x) = range{i1(t,x),i2(t,x), ,in(t,x)}
,in(t,x)}
- enssum
-
Ensemble sum
o(t,x) = sum{i1(t,x),i2(t,x), ,in(t,x)}
,in(t,x)}
- ensmean
-
Ensemble mean
o(t,x) = mean{i1(t,x),i2(t,x), ,in(t,x)}
,in(t,x)}
- ensavg
-
Ensemble average
o(t,x) = avg{i1(t,x),i2(t,x), ,in(t,x)}
,in(t,x)}
- ensstd
-
Ensemble standard deviation
Normalize by n.o(t,x) = std{i1(t,x),i2(t,x),
 ,in(t,x)}
,in(t,x)}
- ensstd1
-
Ensemble standard deviation (n-1)
Normalize by (n-1).o(t,x) = std1{i1(t,x),i2(t,x),
 ,in(t,x)}
,in(t,x)}
- ensvar
-
Ensemble variance
Normalize by n.o(t,x) = var{i1(t,x),i2(t,x),
 ,in(t,x)}
,in(t,x)}
- ensvar1
-
Ensemble variance (n-1)
Normalize by (n-1).o(t,x) = var1{i1(t,x),i2(t,x),
 ,in(t,x)}
,in(t,x)}
- ensskew
-
Ensemble skewness
o(t,x) = skew{i1(t,x),i2(t,x), ,in(t,x)}
,in(t,x)}
- enskurt
-
Ensemble kurtosis
o(t,x) = kurt{i1(t,x),i2(t,x), ,in(t,x)}
,in(t,x)}
- ensmedian
-
Ensemble median
o(t,x) = median{i1(t,x),i2(t,x), ,in(t,x)}
,in(t,x)}
- enspctl
-
Ensemble percentiles
o(t,x) = pth percentile{i1(t,x),i2(t,x), ,in(t,x)}
,in(t,x)}
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
Note
Operators of this module need to open all input files simultaneously. The maximum number of open files depends on the operating system!
Example
To compute the ensemble mean over 6 input files use:
cdo ensmean infile1 infile2 infile3 infile4 infile5 infile6 outfile
Or shorter with filename substitution:
cdo ensmean infile[1-6] outfile
To compute the 50th percentile (median) over 6 input files use:
cdo enspctl,50 infile1 infile2 infile3 infile4 infile5 infile6 outfile
2.8.5 ENSSTAT2 - Statistical values over an ensemble
Synopsis
<operator> obsfile ensfiles outfile
Description
This module computes statistical values over the ensemble of ensfiles using obsfile as a reference. Depending on the operator a ranked Histogram or a roc-curve over all Ensembles ensfiles with reference to obsfile is written to outfile. The date and grid information of a timestep in outfile is the date of the first input file. Thus all input files are required to have the same structure in terms of the gridsize, variable definitions and number of timesteps.
All Operators in this module use obsfile as the reference (for instance an observation) whereas ensfiles are understood as an ensemble consisting of n (where n is the number of ensfiles) members.
The operators ensrkhistspace and ensrkhisttime compute Ranked Histograms. Therefor the vertical axis is utilized as the Histogram axis, which prohibits the use of files containing more than one level. The histogram axis has nensfiles+1 bins with level 0 containing for each grid point the number of observations being smaller as all ensembles and level nensfiles+1 indicating the number of observations being larger than all ensembles.
ensrkhisttime computes a ranked histogram at each timestep reducing each horizontal grid to a 1x1 grid and keeping the time axis as in obsfile. Contrary ensrkhistspace computes a histogram at each grid point keeping the horizontal grid for each variable and reducing the time-axis. The time information is that from the last timestep in obsfile.
Operators
- ensrkhistspace
-
Ranked Histogram averaged over space
- ensrkhisttime
-
Ranked Histogram averaged over time
- ensroc
-
Ensemble Receiver Operating characteristics
Example
To compute a rank histogram over 5 input files ensfile1-ensfile5 given an observation in obsfile use:
cdo ensrkhisttime obsfile ensfile1 ensfile2 ensfile3 ensfile4 ensfile5 outfile
Or shorter with filename substitution:
cdo ensrkhisttime obsfile ensfile[1-5] outfile
2.8.6 ENSVAL - Ensemble validation tools
Synopsis
enscrps rfile infiles outfilebase
ensbrs,x rfile infiles outfilebase
Description
This module computes ensemble validation scores and their decomposition such as the Brier and cumulative ranked probability score (CRPS). The first file is used as a reference it can be a climatology, observation or reanalysis against which the skill of the ensembles given in infiles is measured. Depending on the operator a number of output files is generated each containing the skill score and its decomposition corresponding to the operator. The output is averaged over horizontal fields using appropriate weights for each level and timestep in rfile.
All input files need to have the same structure with the same variables. The date information of a timestep in outfile is the date of the first input file. The output files are named as <outfilebase>.<type>.<filesuffix> where <type> depends on the operator and <filesuffix> is determined from the output file type. There are three output files for operator enscrps and four output files for operator ensbrs.
The CRPS and its decomposition into Reliability and the potential CRPS are calculated by an appropriate averaging over the field members (note, that the CRPS does *not* average linearly). In the three output files <type> has the following meaning: crps for the CRPS, reli for the reliability and crpspot for the potential crps. The relation CRPS = CRPSpot + RELI
holds.
The Brier score of the Ensemble given by infiles with respect to the reference given in rfile and the threshold x is calculated. In the four output files <type> has the following meaning: brs for the Brier score wrt threshold x; brsreli for the Brier score reliability wrt threshold x; brsreso for the Brier score resolution wrt threshold x; brsunct for the Brier score uncertainty wrt threshold x. In analogy to the CRPS the following relation holds: BRS(x) = RELI(x) - RESO(x) + UNCT(x).
The implementation of the decomposition of the CRPS and Brier Score follows Hans Hersbach (2000): Decomposition of the Continuous Ranked Probability Score for Ensemble Prediction Systems, in: Weather and Forecasting (15) pp. 559-570.
The CRPS code decomposition has been verified against the CRAN - ensemble validation package from R. Differences occur when grid-cell area is not uniform as the implementation in R does not account for that.
Operators
- enscrps
-
Ensemble CRPS and decomposition
- ensbrs
-
Ensemble Brier score
Ensemble Brier Score and Decomposition
Example
To compute the field averaged Brier score at x=5 over an ensemble with 5 members ensfile1-5 w.r.t. the reference rfile and write the results to files obase.brs.<suff>, obase.brsreli<suff>, obase.brsreso<suff>, obase.brsunct<suff> where <suff> is determined from the output file type, use
cdo ensbrs,5 rfile ensfile1 ensfile2 ensfile3 ensfile4 ensfile5 obase
or shorter using file name substitution:
cdo ensbrs,5 rfile ensfile[1-5] obase
2.8.7 FLDSTAT - Statistical values over a field
Synopsis
<operator>[,parameter] infile outfile
fldpctl,pn infile outfile
Description
This module computes statistical values of all input fields. A field is a horizontal layer of a data variable. Depending on the chosen operator, the minimum, maximum, range, sum, integral, average, standard deviation, variance, skewness, kurtosis, median or a certain percentile of the field is written to outfile.
Operators
- fldmin
-
Field minimum
For every gridpoint x1,...,xn of the same field it is:
o(t,1) = min{i(t,x′),x1 ≤ x′≤ xn} - fldmax
-
Field maximum
For every gridpoint x1,...,xn of the same field it is:
o(t,1) = max{i(t,x′),x1 ≤ x′≤ xn} - fldrange
-
Field range
For every gridpoint x1,...,xn of the same field it is:
o(t,1) = range{i(t,x′),x1 ≤ x′≤ xn} - fldsum
-
Field sum
For every gridpoint x1,...,xn of the same field it is:
o(t,1) = sum{i(t,x′),x1 ≤ x′≤ xn} - fldint
-
Field integral
For every gridpoint x1,...,xn of the same field it is:
o(t,1) = sum{i(t,x′) * cellarea(x′),x1 ≤ x′≤ xn} - fldmean
-
Field mean
For every gridpoint x1,...,xn of the same field it is:
o(t,1) = mean{i(t,x′),x1 ≤ x′≤ xn}weighted by area weights obtained by the input field.
- fldavg
-
Field average
For every gridpoint x1,...,xn of the same field it is:
o(t,1) = avg{i(t,x′),x1 ≤ x′≤ xn}weighted by area weights obtained by the input field.
- fldstd
-
Field standard deviation
Normalize by n. For every gridpoint x1,...,xn of the same field it is:
o(t,1) = std{i(t,x′),x1 ≤ x′≤ xn}weighted by area weights obtained by the input field.
- fldstd1
-
Field standard deviation (n-1)
Normalize by (n-1). For every gridpoint x1,...,xn of the same field it is:
o(t,1) = std1{i(t,x′),x1 ≤ x′≤ xn}weighted by area weights obtained by the input field.
- fldvar
-
Field variance
Normalize by n. For every gridpoint x1,...,xn of the same field it is:
o(t,1) = var{i(t,x′),x1 ≤ x′≤ xn}weighted by area weights obtained by the input field.
- fldvar1
-
Field variance (n-1)
Normalize by (n-1). For every gridpoint x1,...,xn of the same field it is:
o(t,1) = var1{i(t,x′),x1 ≤ x′≤ xn}weighted by area weights obtained by the input field.
- fldskew
-
Field skewness
For every gridpoint x1,...,xn of the same field it is:
o(t,1) = skew{i(t,x′),x1 ≤ x′≤ xn} - fldkurt
-
Field kurtosis
For every gridpoint x1,...,xn of the same field it is:
o(t,1) = kurt{i(t,x′),x1 ≤ x′≤ xn} - fldmedian
-
Field median
For every gridpoint x1,...,xn of the same field it is:
o(t,1) = median{i(t,x′),x1 ≤ x′≤ xn} - fldcount
-
Field count
Number of non-missing values of the field. - fldpctl
-
Field percentiles
For every gridpoint x1,...,xn of the same field it is:
o(t,1) = pth percentile{i(t,x′),x1 ≤ x′≤ xn}
Parameter
- verbose
-
BOOL print lon/lat coordinates of min/max values
- weights
-
BOOL weights=FALSE disables weighting by grid cell area [default: weights=TRUE]
- pn
-
FLOAT Percentile number in 0, ..., 100
Example
To compute the field mean of all input fields use:
cdo fldmean infile outfile
To compute the 90th percentile of all input fields use:
cdo fldpctl,pn=90 infile outfile
2.8.8 ZONSTAT - Zonal statistics
Synopsis
<operator> infile outfile
zonmean[,zonaldes] infile outfile
zonpctl,p infile outfile
Description
This module computes zonal statistical values of the input fields. Depending on the chosen operator, the zonal minimum, maximum, range, sum, average, standard deviation, variance, skewness, kurtosis, median or a certain percentile of the field is written to outfile. Operators of this module require all variables on the same regular lon/lat grid. Only the zonal mean (zonmean) can be calculated for data on an unstructured grid if the latitude bins are defined with the optional parameter zonaldes.
Operators
- zonmin
-
Zonal minimum
For every latitude the minimum over all longitudes is computed. - zonmax
-
Zonal maximum
For every latitude the maximum over all longitudes is computed. - zonrange
-
Zonal range
For every latitude the range over all longitudes is computed. - zonsum
-
Zonal sum
For every latitude the sum over all longitudes is computed. - zonmean
-
Zonal mean
For every latitude the mean over all longitudes is computed. Use the optional parameter zonaldes for data on an unstructured grid. - zonavg
-
Zonal average
For every latitude the average over all longitudes is computed. - zonstd
-
Zonal standard deviation
For every latitude the standard deviation over all longitudes is computed. Normalize by n. - zonstd1
-
Zonal standard deviation (n-1)
For every latitude the standard deviation over all longitudes is computed. Normalize by (n-1). - zonvar
-
Zonal variance
For every latitude the variance over all longitudes is computed. Normalize by n. - zonvar1
-
Zonal variance (n-1)
For every latitude the variance over all longitudes is computed. Normalize by (n-1). - zonskew
-
Zonal skewness
For every latitude the skewness over all longitudes is computed. - zonkurt
-
Zonal kurtosis
For every latitude the kurtosis over all longitudes is computed. - zonmedian
-
Zonal median
For every latitude the median over all longitudes is computed. - zonpctl
-
Zonal percentiles
For every latitude the pth percentile over all longitudes is computed.
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
- zonaldes
-
STRING Description of the zonal latitude bins needed for data on an unstructured grid. A predefined zonal description is zonal_<DY>. DY is the increment of the latitudes in degrees.
Example
To compute the zonal mean of all input fields use:
cdo zonmean infile outfile
To compute the 50th meridional percentile (median) of all input fields use:
cdo zonpctl,50 infile outfile
2.8.9 MERSTAT - Meridional statistics
Synopsis
<operator> infile outfile
merpctl,p infile outfile
Description
This module computes meridional statistical values of the input fields. Depending on the chosen operator, the meridional minimum, maximum, range, sum, average, standard deviation, variance, skewness, kurtosis, median or a certain percentile of the field is written to outfile. Operators of this module require all variables on the same regular lon/lat grid.
Operators
- mermin
-
Meridional minimum
For every longitude the minimum over all latitudes is computed. - mermax
-
Meridional maximum
For every longitude the maximum over all latitudes is computed. - merrange
-
Meridional range
For every longitude the range over all latitudes is computed. - mersum
-
Meridional sum
For every longitude the sum over all latitudes is computed. - mermean
-
Meridional mean
For every longitude the area weighted mean over all latitudes is computed. - meravg
-
Meridional average
For every longitude the area weighted average over all latitudes is computed. - merstd
-
Meridional standard deviation
For every longitude the standard deviation over all latitudes is computed. Normalize by n. - merstd1
-
Meridional standard deviation (n-1)
For every longitude the standard deviation over all latitudes is computed. Normalize by (n-1). - mervar
-
Meridional variance
For every longitude the variance over all latitudes is computed. Normalize by n. - mervar1
-
Meridional variance (n-1)
For every longitude the variance over all latitudes is computed. Normalize by (n-1). - merskew
-
Meridional skewness
For every longitude the skewness over all latitudes is computed. - merkurt
-
Meridional kurtosis
For every longitude the kurtosis over all latitudes is computed. - mermedian
-
Meridional median
For every longitude the median over all latitudes is computed. - merpctl
-
Meridional percentiles
For every longitude the pth percentile over all latitudes is computed.
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
Example
To compute the meridional mean of all input fields use:
cdo mermean infile outfile
To compute the 50th meridional percentile (median) of all input fields use:
cdo merpctl,50 infile outfile
2.8.10 GRIDBOXSTAT - Statistical values over grid boxes
Synopsis
<operator>,nx,ny infile outfile
Description
This module computes statistical values over surrounding grid boxes. Depending on the chosen operator, the minimum, maximum, range, sum, average, standard deviation, variance, skewness, kurtosis or median of the neighboring grid boxes is written to outfile. All gridbox operators only work on quadrilateral curvilinear grids.
Operators
- gridboxmin
-
Gridbox minimum
Minimum value of the selected grid boxes. - gridboxmax
-
Gridbox maximum
Maximum value of the selected grid boxes. - gridboxrange
-
Gridbox range
Range (max-min value) of the selected grid boxes. - gridboxsum
-
Gridbox sum
Sum of the selected grid boxes. - gridboxmean
-
Gridbox mean
Mean of the selected grid boxes. - gridboxavg
-
Gridbox average
Average of the selected grid boxes. - gridboxstd
-
Gridbox standard deviation
Standard deviation of the selected grid boxes. Normalize by n. - gridboxstd1
-
Gridbox standard deviation (n-1)
Standard deviation of the selected grid boxes. Normalize by (n-1). - gridboxvar
-
Gridbox variance
Variance of the selected grid boxes. Normalize by n. - gridboxvar1
-
Gridbox variance (n-1)
Variance of the selected grid boxes. Normalize by (n-1). - gridboxskew
-
Gridbox skewness
Skewness of the selected grid boxes. - gridboxkurt
-
Gridbox kurtosis
Kurtosis of the selected grid boxes. - gridboxmedian
-
Gridbox median
Median of the selected grid boxes.
Parameter
- nx
-
INTEGER Number of grid boxes in x direction
- ny
-
INTEGER Number of grid boxes in y direction
Example
To compute the mean over 10x10 grid boxes of the input field use:
cdo gridboxmean,10,10 infile outfile
2.8.11 REMAPSTAT - Remaps source points to target cells
Synopsis
<operator>,grid infile outfile
Description
This module maps source points to target cells by calculating a statistical value from the source points. Each target cell contains the statistical value from all source points within that target cell. If there are no source points within a target cell, it gets a missing value. Depending on the chosen operator the minimum, maximum, range, sum, average, variance, standard deviation, skewness, kurtosis or median of source points is computed.
Operators
- remapmin
-
Remap minimum
Minimum value of the source points. - remapmax
-
Remap maximum
Maximum value of the source points. - remaprange
-
Remap range
Range (max-min value) of the source points. - remapsum
-
Remap sum
Sum of the source points. - remapmean
-
Remap mean
Mean of the source points. - remapavg
-
Remap average
Average of the source points. - remapstd
-
Remap standard deviation
Standard deviation of the source points. Normalize by n. - remapstd1
-
Remap standard deviation (n-1)
Standard deviation of the source points. Normalize by (n-1). - remapvar
-
Remap variance
Variance of the source points. Normalize by n. - remapvar1
-
Remap variance (n-1)
Variance of the source points. Normalize by (n-1). - remapskew
-
Remap skewness
Skewness of the source points. - remapkurt
-
Remap kurtosis
Kurtosis of the source points. - remapmedian
-
Remap median
Median of the source points.
Parameter
- grid
-
STRING Target grid description file or name
Example
To compute the mean over source points within the taget cells, use:
cdo remapmean,<targetgrid> infile outfile
If some of the target cells contain missing values, use the Operator setmisstonn to fill these missing values with the nearest neighbor cell:
cdo setmisstonn -remapmean,<targetgrid> infile outfile
2.8.12 VERTSTAT - Vertical statistics
Synopsis
<operator>,weights infile outfile
Description
This module computes statistical values over all levels of the input variables. According to chosen operator the vertical minimum, maximum, range, sum, average, variance or standard deviation is written to outfile.
Operators
- vertmin
-
Vertical minimum
For every gridpoint the minimum over all levels is computed. - vertmax
-
Vertical maximum
For every gridpoint the maximum over all levels is computed. - vertrange
-
Vertical range
For every gridpoint the range over all levels is computed. - vertsum
-
Vertical sum
For every gridpoint the sum over all levels is computed. - vertmean
-
Vertical mean
For every gridpoint the layer weighted mean over all levels is computed. - vertavg
-
Vertical average
For every gridpoint the layer weighted average over all levels is computed. - vertstd
-
Vertical standard deviation
For every gridpoint the standard deviation over all levels is computed. Normalize by n. - vertstd1
-
Vertical standard deviation (n-1)
For every gridpoint the standard deviation over all levels is computed. Normalize by (n-1). - vertvar
-
Vertical variance
For every gridpoint the variance over all levels is computed. Normalize by n. - vertvar1
-
Vertical variance (n-1)
For every gridpoint the variance over all levels is computed. Normalize by (n-1).
Parameter
- weights
-
BOOL weights=FALSE disables weighting by layer thickness [default: weights=TRUE]
Example
To compute the vertical sum of all input variables use:
cdo vertsum infile outfile
2.8.13 TIMSELSTAT - Time range statistics
Synopsis
<operator>,nsets[,noffset[,nskip]] infile outfile
Description
This module computes statistical values for a selected number of timesteps. According to the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of the selected timesteps is written to outfile. The time of outfile is determined by the time in the middle of all contributing timesteps of infile. This can be change with the CDO option --timestat_date <first|middle|last>.
Operators
- timselmin
-
Time selection minimum
For every adjacent sequence t1,...,tn of timesteps of the same selected time range it is:
o(t,x) = min{i(t′,x),t1 ≤ t′≤ tn} - timselmax
-
Time selection maximum
For every adjacent sequence t1,...,tn of timesteps of the same selected time range it is:
o(t,x) = max{i(t′,x),t1 ≤ t′≤ tn} - timselrange
-
Time selection range
For every adjacent sequence t1,...,tn of timesteps of the same selected time range it is:
o(t,x) = range{i(t′,x),t1 ≤ t′≤ tn} - timselsum
-
Time selection sum
For every adjacent sequence t1,...,tn of timesteps of the same selected time range it is:
o(t,x) = sum{i(t′,x),t1 ≤ t′≤ tn} - timselmean
-
Time selection mean
For every adjacent sequence t1,...,tn of timesteps of the same selected time range it is:
o(t,x) = mean{i(t′,x),t1 ≤ t′≤ tn} - timselavg
-
Time selection average
For every adjacent sequence t1,...,tn of timesteps of the same selected time range it is:
o(t,x) = avg{i(t′,x),t1 ≤ t′≤ tn} - timselstd
-
Time selection standard deviation
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same selected time range it is:
o(t,x) = std{i(t′,x),t1 ≤ t′≤ tn} - timselstd1
-
Time selection standard deviation (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same selected time range it is:
o(t,x) = std1{i(t′,x),t1 ≤ t′≤ tn} - timselvar
-
Time selection variance
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same selected time range it is:
o(t,x) = var{i(t′,x),t1 ≤ t′≤ tn} - timselvar1
-
Time selection variance (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same selected time range it is:
o(t,x) = var1{i(t′,x),t1 ≤ t′≤ tn}
Parameter
- nsets
-
INTEGER Number of input timesteps for each output timestep
- noffset
-
INTEGER Number of input timesteps skipped before the first timestep range (optional)
- nskip
-
INTEGER Number of input timesteps skipped between timestep ranges (optional)
Example
Assume an input dataset has monthly means over several years. To compute seasonal means from monthly means the first two month have to be skipped:
cdo timselmean,3,2 infile outfile
2.8.14 TIMSELPCTL - Time range percentile values
Synopsis
timselpctl,p,nsets[,noffset[,nskip]] infile1 infile2 infile3 outfile
Description
This operator computes percentile values over a selected number of timesteps in infile1. The algorithm uses histograms with minimum and maximum bounds given in infile2 and infile3, respectively. The default number of histogram bins is 101. The default can be overridden by setting the environment variable CDO_PCTL_NBINS to a different value. The files infile2 and infile3 should be the result of corresponding timselmin and timselmax operations, respectively. The time of outfile is determined by the time in the middle of all contributing timesteps of infile1. This can be change with the CDO option --timestat_date <first|middle|last>.
For every adjacent sequence t1,...,tn of timesteps of the same selected time range it is:
o(t,x) = pth percentile{i(t′,x),t1 ≤ t′≤ tn}
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
- nsets
-
INTEGER Number of input timesteps for each output timestep
- noffset
-
INTEGER Number of input timesteps skipped before the first timestep range (optional)
- nskip
-
INTEGER Number of input timesteps skipped between timestep ranges (optional)
Environment
- CDO_PCTL_NBINS
-
Sets the number of histogram bins. The default number is 101.
2.8.15 RUNSTAT - Running statistics
Synopsis
<operator>,nts infile outfile
Description
This module computes running statistical values over a selected number of timesteps. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of a selected number of consecutive timesteps read from infile is written to outfile. The time of outfile is determined by the time in the middle of all contributing timesteps of infile. This can be change with the CDO option --timestat_date <first|middle|last>.
Operators
- runmin
-
Running minimum
o(t + (nts - 1)∕2,x) = min{i(t,x),i(t + 1,x),...,i(t + nts - 1,x)} - runmax
-
Running maximum
o(t + (nts - 1)∕2,x) = max{i(t,x),i(t + 1,x),...,i(t + nts - 1,x)} - runrange
-
Running range
o(t + (nts - 1)∕2,x) = range{i(t,x),i(t + 1,x),...,i(t + nts - 1,x)} - runsum
-
Running sum
o(t + (nts - 1)∕2,x) = sum{i(t,x),i(t + 1,x),...,i(t + nts - 1,x)} - runmean
-
Running mean
o(t + (nts - 1)∕2,x) = mean{i(t,x),i(t + 1,x),...,i(t + nts - 1,x)} - runavg
-
Running average
o(t + (nts - 1)∕2,x) = avg{i(t,x),i(t + 1,x),...,i(t + nts - 1,x)} - runstd
-
Running standard deviation
Normalize by n.o(t + (nts - 1)∕2,x) = std{i(t,x),i(t + 1,x),...,i(t + nts - 1,x)}
- runstd1
-
Running standard deviation (n-1)
Normalize by (n-1).o(t + (nts - 1)∕2,x) = std1{i(t,x),i(t + 1,x),...,i(t + nts - 1,x)}
- runvar
-
Running variance
Normalize by n.o(t + (nts - 1)∕2,x) = var{i(t,x),i(t + 1,x),...,i(t + nts - 1,x)}
- runvar1
-
Running variance (n-1)
Normalize by (n-1).o(t + (nts - 1)∕2,x) = var1{i(t,x),i(t + 1,x),...,i(t + nts - 1,x)}
Parameter
- nts
-
INTEGER Number of timesteps
Environment
- CDO_TIMESTAT_DATE
-
Sets the time stamp in outfile to the "first", "middle" or "last" contributing timestep of infile.
Example
To compute the running mean over 9 timesteps use:
cdo runmean,9 infile outfile
2.8.16 RUNPCTL - Running percentile values
Synopsis
runpctl,p,nts infile outfile
Description
This module computes running percentiles over a selected number of timesteps in infile. The time of outfile is determined by the time in the middle of all contributing timesteps of infile. This can be change with the CDO option --timestat_date <first|middle|last>.
o(t + (nts - 1)∕2,x) = pth percentile{i(t,x),i(t + 1,x),...,i(t + nts - 1,x)}
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
- nts
-
INTEGER Number of timesteps
Example
To compute the running 50th percentile (median) over 9 timesteps use:
cdo runpctl,50,9 infile outfile
2.8.17 TIMSTAT - Statistical values over all timesteps
Synopsis
<operator> infile outfile
Description
This module computes statistical values over all timesteps in infile. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of all timesteps read from infile is written to outfile. The time of outfile is determined by the time in the middle of all contributing timesteps of infile. This can be change with the CDO option --timestat_date <first|middle|last>.
Operators
- timmin
-
Time minimum
o(1,x) = min{i(t′,x),t1 ≤ t′≤ tn} - timmax
-
Time maximum
o(1,x) = max{i(t′,x),t1 ≤ t′≤ tn} - timminidx
-
Index of time minimum
o(1,x) = minidx{i(t′,x),t1 ≤ t′≤ tn} - timmaxidx
-
Index of time maximum
o(1,x) = maxidx{i(t′,x),t1 ≤ t′≤ tn} - timrange
-
Time range
o(1,x) = range{i(t′,x),t1 ≤ t′≤ tn} - timsum
-
Time sum
o(1,x) = sum{i(t′,x),t1 ≤ t′≤ tn} - timmean
-
Time mean
o(1,x) = mean{i(t′,x),t1 ≤ t′≤ tn} - timavg
-
Time average
o(1,x) = avg{i(t′,x),t1 ≤ t′≤ tn} - timstd
-
Time standard deviation
Normalize by n. o(1,x) = std{i(t′,x),t1 ≤ t′≤ tn} - timstd1
-
Time standard deviation (n-1)
Normalize by (n-1). o(1,x) = std1{i(t′,x),t1 ≤ t′≤ tn} - timvar
-
Time variance
Normalize by n. o(1,x) = var{i(t′,x),t1 ≤ t′≤ tn} - timvar1
-
Time variance (n-1)
Normalize by (n-1). o(1,x) = var1{i(t′,x),t1 ≤ t′≤ tn}
Example
To compute the mean over all input timesteps use:
cdo timmean infile outfile
2.8.18 TIMPCTL - Percentile values over all timesteps
Synopsis
timpctl,p infile1 infile2 infile3 outfile
Description
This operator computes percentiles over all timesteps in infile1. The algorithm uses histograms with minimum and maximum bounds given in infile2 and infile3, respectively. The default number of histogram bins is 101. The default can be overridden by defining the environment variable CDO_PCTL_NBINS. The files infile2 and infile3 should be the result of corresponding timmin and timmax operations, respectively. The time of outfile is determined by the time in the middle of all contributing timesteps of infile1. This can be change with the CDO option --timestat_date <first|middle|last>.
o(1,x) = pth percentile{i(t′,x),t1 ≤ t′≤ tn}
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
Environment
- CDO_PCTL_NBINS
-
Sets the number of histogram bins. The default number is 101.
Example
To compute the 90th percentile over all input timesteps use:
cdo timmin infile minfile cdo timmax infile maxfile cdo timpctl,90 infile minfile maxfile outfile
Or shorter using operator piping:
cdo timpctl,90 infile -timmin infile -timmax infile outfile
2.8.19 HOURSTAT - Hourly statistics
Synopsis
<operator> infile outfile
Description
This module computes statistical values over timesteps of the same hour. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of timesteps of the same hour is written to outfile. The time of outfile is determined by the time in the middle of all contributing timesteps of infile. This can be change with the CDO option --timestat_date <first|middle|last>.
Operators
- hourmin
-
Hourly minimum
For every adjacent sequence t1,...,tn of timesteps of the same hour it is:
o(t,x) = min{i(t′,x),t1 ≤ t′≤ tn} - hourmax
-
Hourly maximum
For every adjacent sequence t1,...,tn of timesteps of the same hour it is:
o(t,x) = max{i(t′,x),t1 ≤ t′≤ tn} - hourrange
-
Hourly range
For every adjacent sequence t1,...,tn of timesteps of the same hour it is:
o(t,x) = range{i(t′,x),t1 ≤ t′≤ tn} - hoursum
-
Hourly sum
For every adjacent sequence t1,...,tn of timesteps of the same hour it is:
o(t,x) = sum{i(t′,x),t1 ≤ t′≤ tn} - hourmean
-
Hourly mean
For every adjacent sequence t1,...,tn of timesteps of the same hour it is:
o(t,x) = mean{i(t′,x),t1 ≤ t′≤ tn} - houravg
-
Hourly average
For every adjacent sequence t1,...,tn of timesteps of the same hour it is:
o(t,x) = avg{i(t′,x),t1 ≤ t′≤ tn} - hourstd
-
Hourly standard deviation
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same hour it is:
o(t,x) = std{i(t′,x),t1 ≤ t′≤ tn} - hourstd1
-
Hourly standard deviation (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same hour it is:
o(t,x) = std1{i(t′,x),t1 ≤ t′≤ tn} - hourvar
-
Hourly variance
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same hour it is:
o(t,x) = var{i(t′,x),t1 ≤ t′≤ tn} - hourvar1
-
Hourly variance (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same hour it is:
o(t,x) = var1{i(t′,x),t1 ≤ t′≤ tn}
Example
To compute the hourly mean of a time series use:
cdo hourmean infile outfile
2.8.20 HOURPCTL - Hourly percentile values
Synopsis
hourpctl,p infile1 infile2 infile3 outfile
Description
This operator computes percentiles over all timesteps of the same hour in infile1. The algorithm uses histograms with minimum and maximum bounds given in infile2 and infile3, respectively. The default number of histogram bins is 101. The default can be overridden by defining the environment variable CDO_PCTL_NBINS. The files infile2 and infile3 should be the result of corresponding hourmin and hourmax operations, respectively. The time of outfile is determined by the time in the middle of all contributing timesteps of infile1. This can be change with the CDO option --timestat_date <first|middle|last>.
For every adjacent sequence t1,...,tn of timesteps of the same hour it is:
o(t,x) = pth percentile{i(t′,x),t1 ≤ t′≤ tn}
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
Environment
- CDO_PCTL_NBINS
-
Sets the number of histogram bins. The default number is 101.
Example
To compute the hourly 90th percentile of a time series use:
cdo hourmin infile minfile cdo hourmax infile maxfile cdo hourpctl,90 infile minfile maxfile outfile
Or shorter using operator piping:
cdo hourpctl,90 infile -hourmin infile -hourmax infile outfile
2.8.21 DAYSTAT - Daily statistics
Synopsis
<operator>[,parameter] infile outfile
Description
This module computes statistical values over timesteps of the same day. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of timesteps of the same day is written to outfile. The time of outfile is determined by the time in the middle of all contributing timesteps of infile. This can be change with the CDO option --timestat_date <first|middle|last>.
Operators
- daymin
-
Daily minimum
For every adjacent sequence t1,...,tn of timesteps of the same day it is:
o(t,x) = min{i(t′,x),t1 ≤ t′≤ tn} - daymax
-
Daily maximum
For every adjacent sequence t1,...,tn of timesteps of the same day it is:
o(t,x) = max{i(t′,x),t1 ≤ t′≤ tn} - dayrange
-
Daily range
For every adjacent sequence t1,...,tn of timesteps of the same day it is:
o(t,x) = range{i(t′,x),t1 ≤ t′≤ tn} - daysum
-
Daily sum
For every adjacent sequence t1,...,tn of timesteps of the same day it is:
o(t,x) = sum{i(t′,x),t1 ≤ t′≤ tn} - daymean
-
Daily mean
For every adjacent sequence t1,...,tn of timesteps of the same day it is:
o(t,x) = mean{i(t′,x),t1 ≤ t′≤ tn} - dayavg
-
Daily average
For every adjacent sequence t1,...,tn of timesteps of the same day it is:
o(t,x) = avg{i(t′,x),t1 ≤ t′≤ tn} - daystd
-
Daily standard deviation
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same day it is:
o(t,x) = std{i(t′,x),t1 ≤ t′≤ tn} - daystd1
-
Daily standard deviation (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same day it is:
o(t,x) = std1{i(t′,x),t1 ≤ t′≤ tn} - dayvar
-
Daily variance
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same day it is:
o(t,x) = var{i(t′,x),t1 ≤ t′≤ tn} - dayvar1
-
Daily variance (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same day it is:
o(t,x) = var1{i(t′,x),t1 ≤ t′≤ tn}
Parameter
- complete_only
-
BOOL Process the last day only if it is complete
Example
To compute the daily mean of a time series use:
cdo daymean infile outfile
2.8.22 DAYPCTL - Daily percentile values
Synopsis
daypctl,p infile1 infile2 infile3 outfile
Description
This operator computes percentiles over all timesteps of the same day in infile1. The algorithm uses histograms with minimum and maximum bounds given in infile2 and infile3, respectively. The default number of histogram bins is 101. The default can be overridden by defining the environment variable CDO_PCTL_NBINS. The files infile2 and infile3 should be the result of corresponding daymin and daymax operations, respectively. The time of outfile is determined by the time in the middle of all contributing timesteps of infile1. This can be change with the CDO option --timestat_date <first|middle|last>.
For every adjacent sequence t1,...,tn of timesteps of the same day it is:
o(t,x) = pth percentile{i(t′,x),t1 ≤ t′≤ tn}
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
Environment
- CDO_PCTL_NBINS
-
Sets the number of histogram bins. The default number is 101.
Example
To compute the daily 90th percentile of a time series use:
cdo daymin infile minfile cdo daymax infile maxfile cdo daypctl,90 infile minfile maxfile outfile
Or shorter using operator piping:
cdo daypctl,90 infile -daymin infile -daymax infile outfile
2.8.23 MONSTAT - Monthly statistics
Synopsis
<operator>[,parameter] infile outfile
Description
This module computes statistical values over timesteps of the same month. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of timesteps of the same month is written to outfile. The time of outfile is determined by the time in the middle of all contributing timesteps of infile. This can be change with the CDO option --timestat_date <first|middle|last>.
Operators
- monmin
-
Monthly minimum
For every adjacent sequence t1,...,tn of timesteps of the same month it is:
o(t,x) = min{i(t′,x),t1 ≤ t′≤ tn} - monmax
-
Monthly maximum
For every adjacent sequence t1,...,tn of timesteps of the same month it is:
o(t,x) = max{i(t′,x),t1 ≤ t′≤ tn} - monrange
-
Monthly range
For every adjacent sequence t1,...,tn of timesteps of the same month it is:
o(t,x) = range{i(t′,x),t1 ≤ t′≤ tn} - monsum
-
Monthly sum
For every adjacent sequence t1,...,tn of timesteps of the same month it is:
o(t,x) = sum{i(t′,x),t1 ≤ t′≤ tn} - monmean
-
Monthly mean
For every adjacent sequence t1,...,tn of timesteps of the same month it is:
o(t,x) = mean{i(t′,x),t1 ≤ t′≤ tn} - monavg
-
Monthly average
For every adjacent sequence t1,...,tn of timesteps of the same month it is:
o(t,x) = avg{i(t′,x),t1 ≤ t′≤ tn} - monstd
-
Monthly standard deviation
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same month it is:
o(t,x) = std{i(t′,x),t1 ≤ t′≤ tn} - monstd1
-
Monthly standard deviation (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same month it is:
o(t,x) = std1{i(t′,x),t1 ≤ t′≤ tn} - monvar
-
Monthly variance
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same month it is:
o(t,x) = var{i(t′,x),t1 ≤ t′≤ tn} - monvar1
-
Monthly variance (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same month it is:
o(t,x) = var1{i(t′,x),t1 ≤ t′≤ tn}
Parameter
- complete_only
-
BOOL Process the last month only if it is complete
Example
To compute the monthly mean of a time series use:
cdo monmean infile outfile
2.8.24 MONPCTL - Monthly percentile values
Synopsis
monpctl,p infile1 infile2 infile3 outfile
Description
This operator computes percentiles over all timesteps of the same month in infile1. The algorithm uses histograms with minimum and maximum bounds given in infile2 and infile3, respectively. The default number of histogram bins is 101. The default can be overridden by defining the environment variable CDO_PCTL_NBINS. The files infile2 and infile3 should be the result of corresponding monmin and monmax operations, respectively. The time of outfile is determined by the time in the middle of all contributing timesteps of infile1. This can be change with the CDO option --timestat_date <first|middle|last>.
For every adjacent sequence t1,...,tn of timesteps of the same month it is:
o(t,x) = pth percentile{i(t′,x),t1 ≤ t′≤ tn}
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
Environment
- CDO_PCTL_NBINS
-
Sets the number of histogram bins. The default number is 101.
Example
To compute the monthly 90th percentile of a time series use:
cdo monmin infile minfile cdo monmax infile maxfile cdo monpctl,90 infile minfile maxfile outfile
Or shorter using operator piping:
cdo monpctl,90 infile -monmin infile -monmax infile outfile
2.8.25 YEARMONSTAT - Yearly mean from monthly data
Synopsis
yearmonmean infile outfile
Description
This operator computes the yearly mean of a monthly time series. Each month is weighted with the number of days per month. The time of outfile is determined by the time in the middle of all contributing timesteps of infile.
For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = mean{i(t′,x),t1 ≤ t′≤ tn}
Environment
- CDO_TIMESTAT_DATE
-
Sets the date information in outfile to the "first", "middle" or "last" contributing timestep of infile.
Example
To compute the yearly mean of a monthly time series use:
cdo yearmonmean infile outfile
2.8.26 YEARSTAT - Yearly statistics
Synopsis
<operator>[,parameter] infile outfile
Description
This module computes statistical values over timesteps of the same year. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of timesteps of the same year is written to outfile. The time of outfile is determined by the time in the middle of all contributing timesteps of infile. This can be change with the CDO option --timestat_date <first|middle|last>.
Operators
- yearmin
-
Yearly minimum
For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = min{i(t′,x),t1 ≤ t′≤ tn} - yearmax
-
Yearly maximum
For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = max{i(t′,x),t1 ≤ t′≤ tn} - yearminidx
-
Index of yearly minimum
For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = minidx{i(t′,x),t1 ≤ t′≤ tn} - yearmaxidx
-
Index of yearly maximum
For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = maxidx{i(t′,x),t1 ≤ t′≤ tn} - yearrange
-
Yearly range
For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = range{i(t′,x),t1 ≤ t′≤ tn} - yearsum
-
Yearly sum
For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = sum{i(t′,x),t1 ≤ t′≤ tn} - yearmean
-
Yearly mean
For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = mean{i(t′,x),t1 ≤ t′≤ tn} - yearavg
-
Yearly average
For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = avg{i(t′,x),t1 ≤ t′≤ tn} - yearstd
-
Yearly standard deviation
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = std{i(t′,x),t1 ≤ t′≤ tn} - yearstd1
-
Yearly standard deviation (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = std1{i(t′,x),t1 ≤ t′≤ tn} - yearvar
-
Yearly variance
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = var{i(t′,x),t1 ≤ t′≤ tn} - yearvar1
-
Yearly variance (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = var1{i(t′,x),t1 ≤ t′≤ tn}
Parameter
- complete_only
-
BOOL Process the last year only if it is complete
Note
The operators yearmean and yearavg compute only arithmetical means!
Example
To compute the yearly mean of a time series use:
cdo yearmean infile outfile
To compute the yearly mean from the correct weighted monthly mean use:
cdo yearmonmean infile outfile
2.8.27 YEARPCTL - Yearly percentile values
Synopsis
yearpctl,p infile1 infile2 infile3 outfile
Description
This operator computes percentiles over all timesteps of the same year in infile1. The algorithm uses histograms with minimum and maximum bounds given in infile2 and infile3, respectively. The default number of histogram bins is 101. The default can be overridden by defining the environment variable CDO_PCTL_NBINS. The files infile2 and infile3 should be the result of corresponding yearmin and yearmax operations, respectively. The time of outfile is determined by the time in the middle of all contributing timesteps of infile1. This can be change with the CDO option --timestat_date <first|middle|last>.
For every adjacent sequence t1,...,tn of timesteps of the same year it is:
o(t,x) = pth percentile{i(t′,x),t1 ≤ t′≤ tn}
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
Environment
- CDO_PCTL_NBINS
-
Sets the number of histogram bins. The default number is 101.
Example
To compute the yearly 90th percentile of a time series use:
cdo yearmin infile minfile cdo yearmax infile maxfile cdo yearpctl,90 infile minfile maxfile outfile
Or shorter using operator piping:
cdo yearpctl,90 infile -yearmin infile -yearmax infile outfile
2.8.28 SEASSTAT - Seasonal statistics
Synopsis
<operator> infile outfile
Description
This module computes statistical values over timesteps of the same meteorological season. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of timesteps of the same season is written to outfile. The time of outfile is determined by the time in the middle of all contributing timesteps of infile. This can be change with the CDO option --timestat_date <first|middle|last>. Be careful about the first and the last output timestep, they may be incorrect values if the seasons have incomplete timesteps.
Operators
- seasmin
-
Seasonal minimum
For every adjacent sequence t1,...,tn of timesteps of the same season it is:
o(t,x) = min{i(t′,x),t1 ≤ t′≤ tn} - seasmax
-
Seasonal maximum
For every adjacent sequence t1,...,tn of timesteps of the same season it is:
o(t,x) = max{i(t′,x),t1 ≤ t′≤ tn} - seasrange
-
Seasonal range
For every adjacent sequence t1,...,tn of timesteps of the same season it is:
o(t,x) = range{i(t′,x),t1 ≤ t′≤ tn} - seassum
-
Seasonal sum
For every adjacent sequence t1,...,tn of timesteps of the same season it is:
o(t,x) = sum{i(t′,x),t1 ≤ t′≤ tn} - seasmean
-
Seasonal mean
For every adjacent sequence t1,...,tn of timesteps of the same season it is:
o(t,x) = mean{i(t′,x),t1 ≤ t′≤ tn} - seasavg
-
Seasonal average
For every adjacent sequence t1,...,tn of timesteps of the same season it is:
o(t,x) = avg{i(t′,x),t1 ≤ t′≤ tn} - seasstd
-
Seasonal standard deviation
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same season it is:
o(t,x) = std{i(t′,x),t1 ≤ t′≤ tn} - seasstd1
-
Seasonal standard deviation (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same season it is:
o(t,x) = std1{i(t′,x),t1 ≤ t′≤ tn} - seasvar
-
Seasonal variance
Normalize by n. For every adjacent sequence t1,...,tn of timesteps of the same season it is:
o(t,x) = var{i(t′,x),t1 ≤ t′≤ tn} - seasvar1
-
Seasonal variance (n-1)
Normalize by (n-1). For every adjacent sequence t1,...,tn of timesteps of the same season it is:
o(t,x) = var1{i(t′,x),t1 ≤ t′≤ tn}
Example
To compute the seasonal mean of a time series use:
cdo seasmean infile outfile
2.8.29 SEASPCTL - Seasonal percentile values
Synopsis
seaspctl,p infile1 infile2 infile3 outfile
Description
This operator computes percentiles over all timesteps in infile1 of the same season. The algorithm uses histograms with minimum and maximum bounds given in infile2 and infile3, respectively. The default number of histogram bins is 101. The default can be overridden by defining the environment variable CDO_PCTL_NBINS. The files infile2 and infile3 should be the result of corresponding seasmin and seasmax operations, respectively. The time of outfile is determined by the time in the middle of all contributing timesteps of infile1. This can be change with the CDO option --timestat_date <first|middle|last>. Be careful about the first and the last output timestep, they may be incorrect values if the seasons have incomplete timesteps.
For every adjacent sequence t1,...,tn of timesteps of the same season it is:
o(t,x) = pth percentile{i(t′,x),t1 ≤ t′≤ tn}
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
Environment
- CDO_PCTL_NBINS
-
Sets the number of histogram bins. The default number is 101.
Example
To compute the seasonal 90th percentile of a time series use:
cdo seasmin infile minfile cdo seasmax infile maxfile cdo seaspctl,90 infile minfile maxfile outfile
Or shorter using operator piping:
cdo seaspctl,90 infile -seasmin infile -seasmax infile outfile
2.8.30 YHOURSTAT - Multi-year hourly statistics
Synopsis
<operator> infile outfile
Description
This module computes statistical values of each hour and day of year. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of each hour and day of year in infile is written to outfile. The date information in an output field is the date of the last contributing input field.
Operators
- yhourmin
-
Multi-year hourly minimum
o(0001,x) = min{i(t,x),day(i(t)) = 0001} o(8784,x) = min{i(t,x),day(i(t)) = 8784}
o(8784,x) = min{i(t,x),day(i(t)) = 8784} - yhourmax
-
Multi-year hourly maximum
o(0001,x) = max{i(t,x),day(i(t)) = 0001} o(8784,x) = max{i(t,x),day(i(t)) = 8784}
o(8784,x) = max{i(t,x),day(i(t)) = 8784} - yhourrange
-
Multi-year hourly range
o(0001,x) = range{i(t,x),day(i(t)) = 0001} o(8784,x) = range{i(t,x),day(i(t)) = 8784}
o(8784,x) = range{i(t,x),day(i(t)) = 8784} - yhoursum
-
Multi-year hourly sum
o(0001,x) = sum{i(t,x),day(i(t)) = 0001} o(8784,x) = sum{i(t,x),day(i(t)) = 8784}
o(8784,x) = sum{i(t,x),day(i(t)) = 8784} - yhourmean
-
Multi-year hourly mean
o(0001,x) = mean{i(t,x),day(i(t)) = 0001} o(8784,x) = mean{i(t,x),day(i(t)) = 8784}
o(8784,x) = mean{i(t,x),day(i(t)) = 8784} - yhouravg
-
Multi-year hourly average
o(0001,x) = avg{i(t,x),day(i(t)) = 0001} o(8784,x) = avg{i(t,x),day(i(t)) = 8784}
o(8784,x) = avg{i(t,x),day(i(t)) = 8784} - yhourstd
-
Multi-year hourly standard deviation
Normalize by n.o(0001,x) = std{i(t,x),day(i(t)) = 0001} o(8784,x) = std{i(t,x),day(i(t)) = 8784}
o(8784,x) = std{i(t,x),day(i(t)) = 8784} - yhourstd1
-
Multi-year hourly standard deviation (n-1)
Normalize by (n-1).o(0001,x) = std1{i(t,x),day(i(t)) = 0001} o(8784,x) = std1{i(t,x),day(i(t)) = 8784}
o(8784,x) = std1{i(t,x),day(i(t)) = 8784} - yhourvar
-
Multi-year hourly variance
Normalize by n.o(0001,x) = var{i(t,x),day(i(t)) = 0001} o(8784,x) = var{i(t,x),day(i(t)) = 8784}
o(8784,x) = var{i(t,x),day(i(t)) = 8784} - yhourvar1
-
Multi-year hourly variance (n-1)
Normalize by (n-1).o(0001,x) = var1{i(t,x),day(i(t)) = 0001} o(8784,x) = var1{i(t,x),day(i(t)) = 8784}
o(8784,x) = var1{i(t,x),day(i(t)) = 8784}
2.8.31 DHOURSTAT - Multi-day hourly statistics
Synopsis
<operator> infile outfile
Description
This module computes statistical values of each hour of day. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of each hour of day in infile is written to outfile. The date information in an output field is the date of the last contributing input field.
Operators
- dhourmin
-
Multi-day hourly minimum
o(01,x) = min{i(t,x),day(i(t)) = 01} o(24,x) = min{i(t,x),day(i(t)) = 24}
o(24,x) = min{i(t,x),day(i(t)) = 24} - dhourmax
-
Multi-day hourly maximum
o(01,x) = max{i(t,x),day(i(t)) = 01} o(24,x) = max{i(t,x),day(i(t)) = 24}
o(24,x) = max{i(t,x),day(i(t)) = 24} - dhourrange
-
Multi-day hourly range
o(01,x) = range{i(t,x),day(i(t)) = 01} o(24,x) = range{i(t,x),day(i(t)) = 24}
o(24,x) = range{i(t,x),day(i(t)) = 24} - dhoursum
-
Multi-day hourly sum
o(01,x) = sum{i(t,x),day(i(t)) = 01} o(24,x) = sum{i(t,x),day(i(t)) = 24}
o(24,x) = sum{i(t,x),day(i(t)) = 24} - dhourmean
-
Multi-day hourly mean
o(01,x) = mean{i(t,x),day(i(t)) = 01} o(24,x) = mean{i(t,x),day(i(t)) = 24}
o(24,x) = mean{i(t,x),day(i(t)) = 24} - dhouravg
-
Multi-day hourly average
o(01,x) = avg{i(t,x),day(i(t)) = 01} o(24,x) = avg{i(t,x),day(i(t)) = 24}
o(24,x) = avg{i(t,x),day(i(t)) = 24} - dhourstd
-
Multi-day hourly standard deviation
Normalize by n.o(01,x) = std{i(t,x),day(i(t)) = 01} o(24,x) = std{i(t,x),day(i(t)) = 24}
o(24,x) = std{i(t,x),day(i(t)) = 24} - dhourstd1
-
Multi-day hourly standard deviation (n-1)
Normalize by (n-1).o(01,x) = std1{i(t,x),day(i(t)) = 01} o(24,x) = std1{i(t,x),day(i(t)) = 24}
o(24,x) = std1{i(t,x),day(i(t)) = 24} - dhourvar
-
Multi-day hourly variance
Normalize by n.o(01,x) = var{i(t,x),day(i(t)) = 01} o(24,x) = var{i(t,x),day(i(t)) = 24}
o(24,x) = var{i(t,x),day(i(t)) = 24} - dhourvar1
-
Multi-day hourly variance (n-1)
Normalize by (n-1).o(01,x) = var1{i(t,x),day(i(t)) = 01} o(24,x) = var1{i(t,x),day(i(t)) = 24}
o(24,x) = var1{i(t,x),day(i(t)) = 24}
2.8.32 DMINUTESTAT - Multi-day by the minute statistics
Synopsis
<operator> infile outfile
Description
This module computes statistical values of each minute of day. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of each minute of day in infile is written to outfile. The date information in an output field is the date of the last contributing input field.
Operators
- dminutemin
-
Multi-day by the minute minimum
o(01,x) = min{i(t,x),day(i(t)) = 01} o(1440,x) = min{i(t,x),day(i(t)) = 1440}
o(1440,x) = min{i(t,x),day(i(t)) = 1440} - dminutemax
-
Multi-day by the minute maximum
o(01,x) = max{i(t,x),day(i(t)) = 01} o(1440,x) = max{i(t,x),day(i(t)) = 1440}
o(1440,x) = max{i(t,x),day(i(t)) = 1440} - dminuterange
-
Multi-day by the minute range
o(01,x) = range{i(t,x),day(i(t)) = 01} o(1440,x) = range{i(t,x),day(i(t)) = 1440}
o(1440,x) = range{i(t,x),day(i(t)) = 1440} - dminutesum
-
Multi-day by the minute sum
o(01,x) = sum{i(t,x),day(i(t)) = 01} o(1440,x) = sum{i(t,x),day(i(t)) = 1440}
o(1440,x) = sum{i(t,x),day(i(t)) = 1440} - dminutemean
-
Multi-day by the minute mean
o(01,x) = mean{i(t,x),day(i(t)) = 01} o(1440,x) = mean{i(t,x),day(i(t)) = 1440}
o(1440,x) = mean{i(t,x),day(i(t)) = 1440} - dminuteavg
-
Multi-day by the minute average
o(01,x) = avg{i(t,x),day(i(t)) = 01} o(1440,x) = avg{i(t,x),day(i(t)) = 1440}
o(1440,x) = avg{i(t,x),day(i(t)) = 1440} - dminutestd
-
Multi-day by the minute standard deviation
Normalize by n.o(01,x) = std{i(t,x),day(i(t)) = 01} o(1440,x) = std{i(t,x),day(i(t)) = 1440}
o(1440,x) = std{i(t,x),day(i(t)) = 1440} - dminutestd1
-
Multi-day by the minute standard deviation (n-1)
Normalize by (n-1).o(01,x) = std1{i(t,x),day(i(t)) = 01} o(1440,x) = std1{i(t,x),day(i(t)) = 1440}
o(1440,x) = std1{i(t,x),day(i(t)) = 1440} - dminutevar
-
Multi-day by the minute variance
Normalize by n.o(01,x) = var{i(t,x),day(i(t)) = 01} o(1440,x) = var{i(t,x),day(i(t)) = 1440}
o(1440,x) = var{i(t,x),day(i(t)) = 1440} - dminutevar1
-
Multi-day by the minute variance (n-1)
Normalize by (n-1).o(01,x) = var1{i(t,x),day(i(t)) = 01} o(1440,x) = var1{i(t,x),day(i(t)) = 1440}
o(1440,x) = var1{i(t,x),day(i(t)) = 1440}
2.8.33 YDAYSTAT - Multi-year daily statistics
Synopsis
<operator> infile outfile
Description
This module computes statistical values of each day of year. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of each day of year in infile is written to outfile. The date information in an output field is the date of the last contributing input field.
Operators
- ydaymin
-
Multi-year daily minimum
o(001,x) = min{i(t,x),day(i(t)) = 001} o(366,x) = min{i(t,x),day(i(t)) = 366}
o(366,x) = min{i(t,x),day(i(t)) = 366} - ydaymax
-
Multi-year daily maximum
o(001,x) = max{i(t,x),day(i(t)) = 001} o(366,x) = max{i(t,x),day(i(t)) = 366}
o(366,x) = max{i(t,x),day(i(t)) = 366} - ydayrange
-
Multi-year daily range
o(001,x) = range{i(t,x),day(i(t)) = 001} o(366,x) = range{i(t,x),day(i(t)) = 366}
o(366,x) = range{i(t,x),day(i(t)) = 366} - ydaysum
-
Multi-year daily sum
o(001,x) = sum{i(t,x),day(i(t)) = 001} o(366,x) = sum{i(t,x),day(i(t)) = 366}
o(366,x) = sum{i(t,x),day(i(t)) = 366} - ydaymean
-
Multi-year daily mean
o(001,x) = mean{i(t,x),day(i(t)) = 001} o(366,x) = mean{i(t,x),day(i(t)) = 366}
o(366,x) = mean{i(t,x),day(i(t)) = 366} - ydayavg
-
Multi-year daily average
o(001,x) = avg{i(t,x),day(i(t)) = 001} o(366,x) = avg{i(t,x),day(i(t)) = 366}
o(366,x) = avg{i(t,x),day(i(t)) = 366} - ydaystd
-
Multi-year daily standard deviation
Normalize by n.o(001,x) = std{i(t,x),day(i(t)) = 001} o(366,x) = std{i(t,x),day(i(t)) = 366}
o(366,x) = std{i(t,x),day(i(t)) = 366} - ydaystd1
-
Multi-year daily standard deviation (n-1)
Normalize by (n-1).o(001,x) = std1{i(t,x),day(i(t)) = 001} o(366,x) = std1{i(t,x),day(i(t)) = 366}
o(366,x) = std1{i(t,x),day(i(t)) = 366} - ydayvar
-
Multi-year daily variance
Normalize by n.o(001,x) = var{i(t,x),day(i(t)) = 001} o(366,x) = var{i(t,x),day(i(t)) = 366}
o(366,x) = var{i(t,x),day(i(t)) = 366} - ydayvar1
-
Multi-year daily variance (n-1)
Normalize by (n-1).o(001,x) = var1{i(t,x),day(i(t)) = 001} o(366,x) = var1{i(t,x),day(i(t)) = 366}
o(366,x) = var1{i(t,x),day(i(t)) = 366}
Example
To compute the daily mean over all input years use:
cdo ydaymean infile outfile
2.8.34 YDAYPCTL - Multi-year daily percentile values
Synopsis
ydaypctl,p infile1 infile2 infile3 outfile
Description
This operator writes a certain percentile of each day of year in infile1 to outfile. The algorithm uses histograms with minimum and maximum bounds given in infile2 and infile3, respectively. The default number of histogram bins is 101. The default can be overridden by setting the environment variable CDO_PCTL_NBINS to a different value. The files infile2 and infile3 should be the result of corresponding ydaymin and ydaymax operations, respectively. The date information in an output field is the date of the last contributing input field.
o(001,x) = pth percentile{i(t,x),day(i(t)) = 001} |
 |
o(366,x) = pth percentile{i(t,x),day(i(t)) = 366} |
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
Environment
- CDO_PCTL_NBINS
-
Sets the number of histogram bins. The default number is 101.
Example
To compute the daily 90th percentile over all input years use:
cdo ydaymin infile minfile cdo ydaymax infile maxfile cdo ydaypctl,90 infile minfile maxfile outfile
Or shorter using operator piping:
cdo ydaypctl,90 infile -ydaymin infile -ydaymax infile outfile
2.8.35 YMONSTAT - Multi-year monthly statistics
Synopsis
<operator> infile outfile
Description
This module computes statistical values of each month of year. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of each month of year in infile is written to outfile. The date information in an output field is the date of the last contributing input field. This can be change with the CDO option --timestat_date <first|middle|last>.
Operators
- ymonmin
-
Multi-year monthly minimum
o(01,x) = min{i(t,x),month(i(t)) = 01} o(12,x) = min{i(t,x),month(i(t)) = 12}
o(12,x) = min{i(t,x),month(i(t)) = 12} - ymonmax
-
Multi-year monthly maximum
o(01,x) = max{i(t,x),month(i(t)) = 01} o(12,x) = max{i(t,x),month(i(t)) = 12}
o(12,x) = max{i(t,x),month(i(t)) = 12} - ymonrange
-
Multi-year monthly range
o(01,x) = range{i(t,x),month(i(t)) = 01} o(12,x) = range{i(t,x),month(i(t)) = 12}
o(12,x) = range{i(t,x),month(i(t)) = 12} - ymonsum
-
Multi-year monthly sum
o(01,x) = sum{i(t,x),month(i(t)) = 01} o(12,x) = sum{i(t,x),month(i(t)) = 12}
o(12,x) = sum{i(t,x),month(i(t)) = 12} - ymonmean
-
Multi-year monthly mean
o(01,x) = mean{i(t,x),month(i(t)) = 01} o(12,x) = mean{i(t,x),month(i(t)) = 12}
o(12,x) = mean{i(t,x),month(i(t)) = 12} - ymonavg
-
Multi-year monthly average
o(01,x) = avg{i(t,x),month(i(t)) = 01} o(12,x) = avg{i(t,x),month(i(t)) = 12}
o(12,x) = avg{i(t,x),month(i(t)) = 12} - ymonstd
-
Multi-year monthly standard deviation
Normalize by n.o(01,x) = std{i(t,x),month(i(t)) = 01} o(12,x) = std{i(t,x),month(i(t)) = 12}
o(12,x) = std{i(t,x),month(i(t)) = 12} - ymonstd1
-
Multi-year monthly standard deviation (n-1)
Normalize by (n-1).o(01,x) = std1{i(t,x),month(i(t)) = 01} o(12,x) = std1{i(t,x),month(i(t)) = 12}
o(12,x) = std1{i(t,x),month(i(t)) = 12} - ymonvar
-
Multi-year monthly variance
Normalize by n.o(01,x) = var{i(t,x),month(i(t)) = 01} o(12,x) = var{i(t,x),month(i(t)) = 12}
o(12,x) = var{i(t,x),month(i(t)) = 12} - ymonvar1
-
Multi-year monthly variance (n-1)
Normalize by (n-1).o(01,x) = var1{i(t,x),month(i(t)) = 01} o(12,x) = var1{i(t,x),month(i(t)) = 12}
o(12,x) = var1{i(t,x),month(i(t)) = 12}
Example
To compute the monthly mean over all input years use:
cdo ymonmean infile outfile
2.8.36 YMONPCTL - Multi-year monthly percentile values
Synopsis
ymonpctl,p infile1 infile2 infile3 outfile
Description
This operator writes a certain percentile of each month of year in infile1 to outfile. The algorithm uses histograms with minimum and maximum bounds given in infile2 and infile3, respectively. The default number of histogram bins is 101. The default can be overridden by setting the environment variable CDO_PCTL_NBINS to a different value. The files infile2 and infile3 should be the result of corresponding ymonmin and ymonmax operations, respectively. The date information in an output field is the date of the last contributing input field.
o(01,x) = pth percentile{i(t,x),month(i(t)) = 01} |
 |
o(12,x) = pth percentile{i(t,x),month(i(t)) = 12} |
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
Environment
- CDO_PCTL_NBINS
-
Sets the number of histogram bins. The default number is 101.
Example
To compute the monthly 90th percentile over all input years use:
cdo ymonmin infile minfile cdo ymonmax infile maxfile cdo ymonpctl,90 infile minfile maxfile outfile
Or shorter using operator piping:
cdo ymonpctl,90 infile -ymonmin infile -ymonmax infile outfile
2.8.37 YSEASSTAT - Multi-year seasonal statistics
Synopsis
<operator> infile outfile
Description
This module computes statistical values of each season. Depending on the chosen operator the minimum, maximum, range, sum, average, variance or standard deviation of each season in infile is written to outfile. The date information in an output field is the date of the last contributing input field.
Operators
- yseasmin
-
Multi-year seasonal minimum
o(1,x) = min{i(t,x),month(i(t)) = 12, 01, 02}o(2,x) = min{i(t,x),month(i(t)) = 03, 04, 05}o(3,x) = min{i(t,x),month(i(t)) = 06, 07, 08}o(4,x) = min{i(t,x),month(i(t)) = 09, 10, 11} - yseasmax
-
Multi-year seasonal maximum
o(1,x) = max{i(t,x),month(i(t)) = 12, 01, 02}o(2,x) = max{i(t,x),month(i(t)) = 03, 04, 05}o(3,x) = max{i(t,x),month(i(t)) = 06, 07, 08}o(4,x) = max{i(t,x),month(i(t)) = 09, 10, 11} - yseasrange
-
Multi-year seasonal range
o(1,x) = range{i(t,x),month(i(t)) = 12, 01, 02}o(2,x) = range{i(t,x),month(i(t)) = 03, 04, 05}o(3,x) = range{i(t,x),month(i(t)) = 06, 07, 08}o(4,x) = range{i(t,x),month(i(t)) = 09, 10, 11} - yseassum
-
Multi-year seasonal sum
o(1,x) = sum{i(t,x),month(i(t)) = 12, 01, 02}o(2,x) = sum{i(t,x),month(i(t)) = 03, 04, 05}o(3,x) = sum{i(t,x),month(i(t)) = 06, 07, 08}o(4,x) = sum{i(t,x),month(i(t)) = 09, 10, 11} - yseasmean
-
Multi-year seasonal mean
o(1,x) = mean{i(t,x),month(i(t)) = 12, 01, 02}o(2,x) = mean{i(t,x),month(i(t)) = 03, 04, 05}o(3,x) = mean{i(t,x),month(i(t)) = 06, 07, 08}o(4,x) = mean{i(t,x),month(i(t)) = 09, 10, 11} - yseasavg
-
Multi-year seasonal average
o(1,x) = avg{i(t,x),month(i(t)) = 12, 01, 02}o(2,x) = avg{i(t,x),month(i(t)) = 03, 04, 05}o(3,x) = avg{i(t,x),month(i(t)) = 06, 07, 08}o(4,x) = avg{i(t,x),month(i(t)) = 09, 10, 11} - yseasstd
-
Multi-year seasonal standard deviation
o(1,x) = std{i(t,x),month(i(t)) = 12, 01, 02}o(2,x) = std{i(t,x),month(i(t)) = 03, 04, 05}o(3,x) = std{i(t,x),month(i(t)) = 06, 07, 08}o(4,x) = std{i(t,x),month(i(t)) = 09, 10, 11} - yseasstd1
-
Multi-year seasonal standard deviation (n-1)
o(1,x) = std1{i(t,x),month(i(t)) = 12, 01, 02}o(2,x) = std1{i(t,x),month(i(t)) = 03, 04, 05}o(3,x) = std1{i(t,x),month(i(t)) = 06, 07, 08}o(4,x) = std1{i(t,x),month(i(t)) = 09, 10, 11} - yseasvar
-
Multi-year seasonal variance
o(1,x) = var{i(t,x),month(i(t)) = 12, 01, 02}o(2,x) = var{i(t,x),month(i(t)) = 03, 04, 05}o(3,x) = var{i(t,x),month(i(t)) = 06, 07, 08}o(4,x) = var{i(t,x),month(i(t)) = 09, 10, 11} - yseasvar1
-
Multi-year seasonal variance (n-1)
o(1,x) = var1{i(t,x),month(i(t)) = 12, 01, 02}o(2,x) = var1{i(t,x),month(i(t)) = 03, 04, 05}o(3,x) = var1{i(t,x),month(i(t)) = 06, 07, 08}o(4,x) = var1{i(t,x),month(i(t)) = 09, 10, 11}
Example
To compute the seasonal mean over all input years use:
cdo yseasmean infile outfile
2.8.38 YSEASPCTL - Multi-year seasonal percentile values
Synopsis
yseaspctl,p infile1 infile2 infile3 outfile
Description
This operator writes a certain percentile of each season in infile1 to outfile. The algorithm uses histograms with minimum and maximum bounds given in infile2 and infile3, respectively. The default number of histogram bins is 101. The default can be overridden by setting the environment variable CDO_PCTL_NBINS to a different value. The files infile2 and infile3 should be the result of corresponding yseasmin and yseasmax operations, respectively. The date information in an output field is the date of the last contributing input field.
o(1,x) = pth percentile{i(t,x),month(i(t)) = 12, 01, 02} |
o(2,x) = pth percentile{i(t,x),month(i(t)) = 03, 04, 05} |
o(3,x) = pth percentile{i(t,x),month(i(t)) = 06, 07, 08} |
o(4,x) = pth percentile{i(t,x),month(i(t)) = 09, 10, 11} |
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
Environment
- CDO_PCTL_NBINS
-
Sets the number of histogram bins. The default number is 101.
Example
To compute the seasonal 90th percentile over all input years use:
cdo yseasmin infile minfile cdo yseasmax infile maxfile cdo yseaspctl,90 infile minfile maxfile outfile
Or shorter using operator piping:
cdo yseaspctl,90 infile -yseasmin infile -yseasmax infile outfile
2.8.39 YDRUNSTAT - Multi-year daily running statistics
Synopsis
<operator>,nts[,rm=c] infile outfile
Description
This module writes running statistical values for each day of year in infile to outfile. Depending on the chosen operator, the minimum, maximum, sum, average, variance or standard deviation of all timesteps in running windows of which the medium timestep corresponds to a certain day of year is computed. The date information in an output field is the date of the timestep in the middle of the last contributing running window. Note that the operator have to be applied to a continuous time series of daily measurements in order to yield physically meaningful results. Also note that the output time series begins (nts-1)/2 timesteps after the first timestep of the input time series and ends (nts-1)/2 timesteps before the last one. For input data which are complete but not continuous, such as time series of daily measurements for the same month or season within different years, the operator yields physically meaningful results only if the input time series does include the (nts-1)/2 days before and after each period of interest.
Operators
- ydrunmin
-
Multi-year daily running minimum
o(001,x) = min{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 001}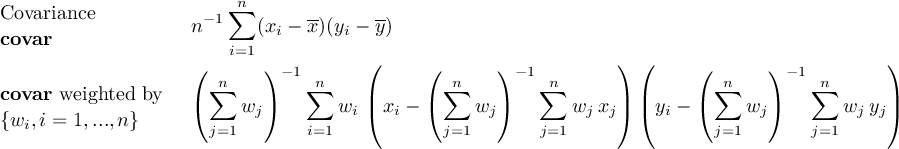 o(366,x) = min{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366}
o(366,x) = min{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366} - ydrunmax
-
Multi-year daily running maximum
o(001,x) = max{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 001}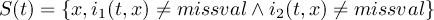 o(366,x) = max{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366}
o(366,x) = max{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366} - ydrunsum
-
Multi-year daily running sum
o(001,x) = sum{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 001}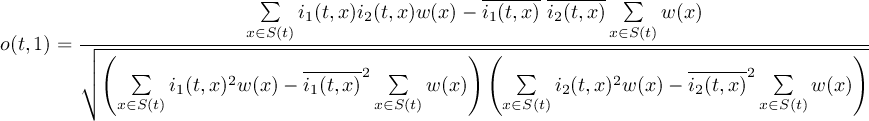 o(366,x) = sum{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366}
o(366,x) = sum{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366} - ydrunmean
-
Multi-year daily running mean
o(001,x) = mean{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 001}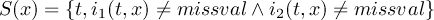 o(366,x) = mean{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366}
o(366,x) = mean{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366} - ydrunavg
-
Multi-year daily running average
o(001,x) = avg{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 001}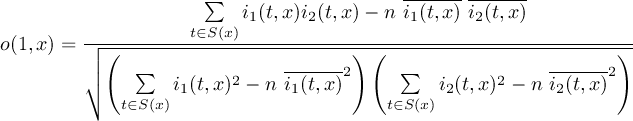 o(366,x) = avg{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366}
o(366,x) = avg{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366} - ydrunstd
-
Multi-year daily running standard deviation
Normalize by n.o(001,x) = std{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 001}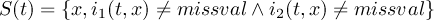 o(366,x) = std{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366}
o(366,x) = std{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366} - ydrunstd1
-
Multi-year daily running standard deviation (n-1)
Normalize by (n-1).o(001,x) = std1{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 001}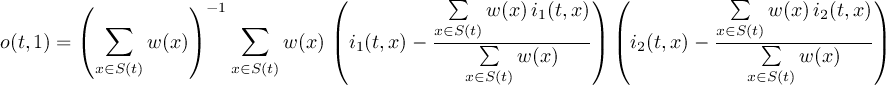 o(366,x) = std1{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366}
o(366,x) = std1{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366} - ydrunvar
-
Multi-year daily running variance
Normalize by n.o(001,x) = var{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 001}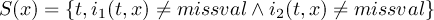 o(366,x) = var{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366}
o(366,x) = var{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366} - ydrunvar1
-
Multi-year daily running variance (n-1)
Normalize by (n-1).o(001,x) = var1{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 001}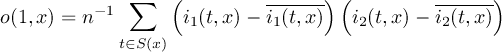 o(366,x) = var1{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366}
o(366,x) = var1{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366}
Parameter
- nts
-
INTEGER Number of timesteps
- rm=c
-
STRING Read method circular
Example
Assume the input data provide a continuous time series of daily measurements. To compute the running multi-year daily mean over all input timesteps for a running window of five days use:
cdo ydrunmean,5 infile outfile
Note that except for the standard deviation the results of the operators in this module are equivalent to a composition of corresponding operators from the YDAYSTAT and RUNSTAT modules. For instance, the above command yields the same result as:
cdo ydaymean -runmean,5 infile outfile
2.8.40 YDRUNPCTL - Multi-year daily running percentile values
Synopsis
ydrunpctl,p,nts[,rm=c[,pm=r8]] infile1 infile2 infile3 outfile
Description
This operator writes running percentile values for each day of year in infile1 to outfile. A certain percentile is computed for all timesteps in running windows of which the medium timestep corresponds to a certain day of year. The algorithm uses histograms with minimum and maximum bounds given in infile2 and infile3, respectively. The default number of histogram bins is 101. The default can be overridden by setting the environment variable CDO_PCTL_NBINS to a different value. The files infile2 and infile3 should be the result of corresponding ydrunmin and ydrunmax operations, respectively. The date information in an output field is the date of the timestep in the middle of the last contributing running window. Note that the operator have to be applied to a continuous time series of daily measurements in order to yield physically meaningful results. Also note that the output time series begins (nts-1)/2 timesteps after the first timestep of the input time series and ends (nts-1)/2 timesteps before the last. For input data which are complete but not continuous, such as time series of daily measurements for the same month or season within different years, the operator only yields physically meaningful results if the input time series does include the (nts-1)/2 days before and after each period of interest.
o(001,x) = pth percentile{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 001} |
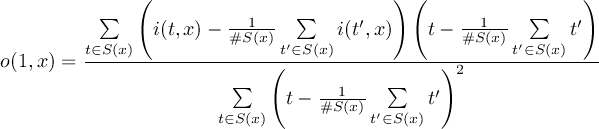 |
o(366,x) = pth percentile{i(t,x),i(t + 1,x),...,i(t + nts - 1,x);day[(i(t + (nts - 1)∕2)] = 366} |
Parameter
- p
-
FLOAT Percentile number in 0, ..., 100
- nts
-
INTEGER Number of timesteps
- rm=c
-
STRING Read method circular
- pm=r8
-
STRING Percentile method rtype8
Environment
- CDO_PCTL_NBINS
-
Sets the number of histogram bins. The default number is 101.
Example
Assume the input data provide a continuous time series of daily measurements. To compute the running multi-year daily 90th percentile over all input timesteps for a running window of five days use:
cdo ydrunmin,5 infile minfile cdo ydrunmax,5 infile maxfile cdo ydrunpctl,90,5 infile minfile maxfile outfile
Or shorter using operator piping:
cdo ydrunpctl,90,5 infile -ydrunmin infile -ydrunmax infile outfile
2.9 Correlation and co.
This sections contains modules for correlation and co. in grid space and over time.
In this section the abbreviations as in the following table are used:
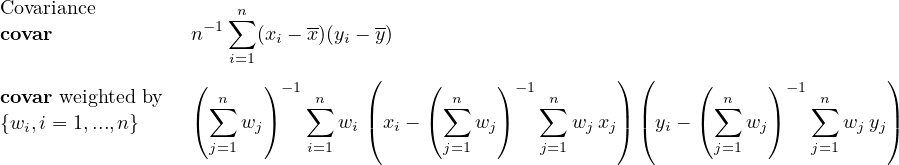
| fldcor | Correlation in grid space |
| timcor | Correlation over time |
| fldcovar | Covariance in grid space |
| timcovar | Covariance over time |
2.9.1 FLDCOR - Correlation in grid space
Synopsis
fldcor infile1 infile2 outfile
Description
The correlation coefficient is a quantity that gives the quality of a least squares fitting to the original data. This operator correlates all gridpoints of two fields for each timestep. With
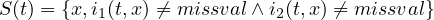
it is
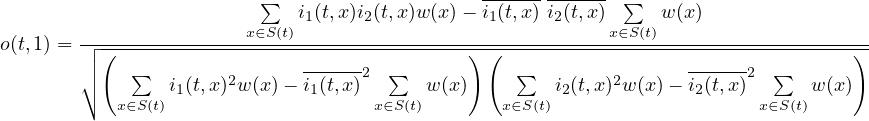
where w(x) are the area weights obtained by the input streams. For every timestep t only those field elements x belong to the sample, which have i1(t,x)≠missval and i2(t,x)≠missval.
2.9.2 TIMCOR - Correlation over time
Synopsis
timcor infile1 infile2 outfile
Description
The correlation coefficient is a quantity that gives the quality of a least squares fitting to the original data. This operator correlates each gridpoint of two fields over all timesteps. If there is only one input field, the p-value (probability value) is also written out. With
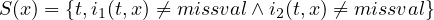
it is
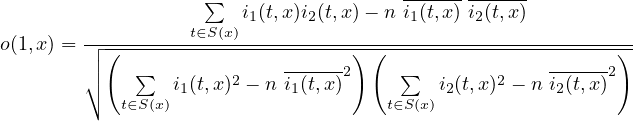
For every gridpoint x only those timesteps t belong to the sample, which have i1(t,x)≠missval and i2(t,x)≠missval.
2.9.3 FLDCOVAR - Covariance in grid space
Synopsis
fldcovar infile1 infile2 outfile
Description
This operator calculates the covariance of two fields over all gridpoints for each timestep. With
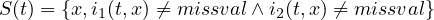
it is
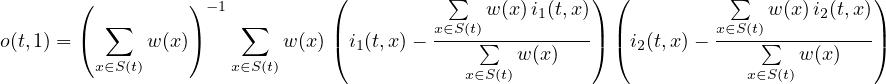
where w(x) are the area weights obtained by the input streams. For every timestep t only those field elements x belong to the sample, which have i1(t,x)≠missval and i2(t,x)≠missval.
2.9.4 TIMCOVAR - Covariance over time
Synopsis
timcovar infile1 infile2 outfile
Description
This operator calculates the covariance of two fields at each gridpoint over all timesteps. With
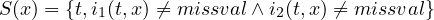
it is
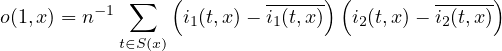
For every gridpoint x only those timesteps t belong to the sample, which have i1(t,x)≠missval and i2(t,x)≠missval.
2.10 Regression
This sections contains modules for linear regression of time series.
Here is a short overview of all operators in this section:
| regres | Regression |
| detrend | Detrend |
| trend | Trend |
| addtrend | Add trend |
| subtrend | Subtract trend |
2.10.1 REGRES - Regression
Synopsis
regres[,equal] infile outfile
Description
The values of the input file infile are assumed to be distributed as N(a + bt,σ2) with unknown a, b and σ2. This operator estimates the parameter b. For every field element x only those timesteps t belong to the sample S(x), which have i(t,x)≠miss. It is
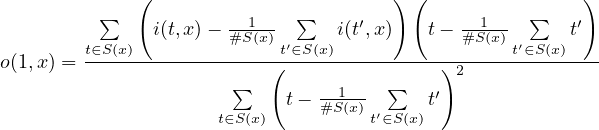
It is assumed that all timesteps are equidistant, if this is not the case set the parameter equal=false.
Parameter
- equal
-
BOOL Set to false for unequal distributed timesteps (default: true)
2.10.2 DETREND - Detrend time series
Synopsis
detrend[,equal] infile outfile
Description
Every time series in infile is linearly detrended. For every field element x only those timesteps t belong to the sample S, which have i(t,x)≠miss. It is assumed that all timesteps are equidistant, if this is not the case set the parameter equal=false. With
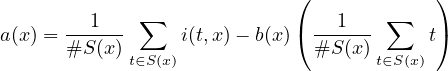
and
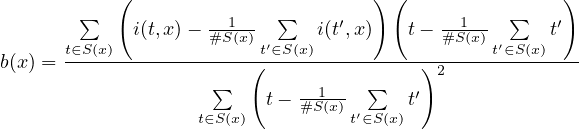
it is
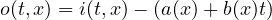
Parameter
- equal
-
BOOL Set to false for unequal distributed timesteps (default: true)
Note
This operator has to keep the fields of all timesteps concurrently in the memory. If not enough memory is available use the operators trend and subtrend.
Example
To detrend the data in infile and to store the detrended data in outfile use:
cdo detrend infile outfile
2.10.3 TREND - Trend of time series
Synopsis
trend[,equal] infile outfile1 outfile2
Description
The values of the input file infile are assumed to be distributed as N(a + bt,σ2) with unknown a, b and σ2. This operator estimates the parameter a and b. For every field element x only those timesteps t belong to the sample S(x), which have i(t,x)≠miss. It is
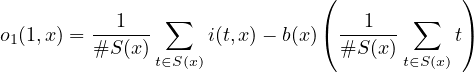
and
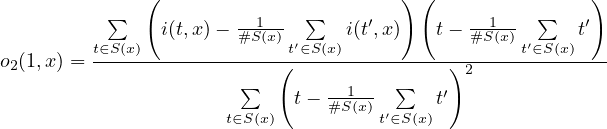
Thus the estimation for a is stored in outfile1 and that for b is stored in outfile2. To subtract the trend from the data see operator subtrend. It is assumed that all timesteps are equidistant, if this is not the case set the parameter equal=false.
Parameter
- equal
-
BOOL Set to false for unequal distributed timesteps (default: true)
2.10.4 TRENDARITH - Add or subtract a trend
Synopsis
<operator>[,equal] infile1 infile2 infile3 outfile
Description
This module is for adding or subtracting a trend computed by the operator trend.
Operators
- addtrend
-
Add trend
It is
where t is the timesteps.
- subtrend
-
Subtract trend
It is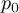
where t is the timesteps.
Parameter
- equal
-
BOOL Set to false for unequal distributed timesteps (default: true)
Example
The typical call for detrending the data in infile and storing the detrended data in outfile is:
cdo trend infile afile bfile cdo subtrend infile afile bfile outfile
The result is identical to a call of the operator detrend:
cdo detrend infile outfile
2.11 Eof
This section contains modules to compute Empirical Orthogonal Functions and - once they are computed - their
principal coefficients.
An introduction to the theory of principal component analysis as applied here can be found in:
Principal Component Analysis in Meteorology and Oceanography [Preisendorfer]
Details about calculation in the time- and spatial spaces are found in:
Statistical Analysis in Climate Research [vonStorch]
EOFs are defined as the eigen values of the scatter matrix (covariance matrix) of the data. For the sake of simplicity, samples are regarded as time series of anomalies
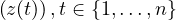
of (column-) vectors 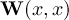 with
with 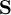 entries (where
entries (where 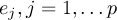 is the gridsize). Thus, using the fact, that
is the gridsize). Thus, using the fact, that 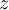 are
anomalies, i.e.
are
anomalies, i.e.
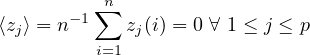
the scatter matrix  can be written as
can be written as
![∑n [√--- ][√--- ]T
S = Wz (t) Wz (t)
t=1](cdo147x.png)
where 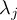 is the diagonal matrix containing the area weight of cell
is the diagonal matrix containing the area weight of cell 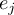 in
in 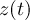 at
at 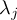 .
.
The matrix 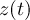 has a set of orthonormal eigenvectors
has a set of orthonormal eigenvectors 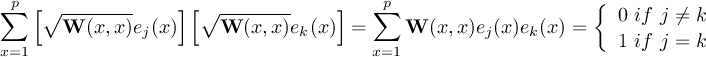 , which are called empirical orthogonal
functions (EOFs) of the sample
, which are called empirical orthogonal
functions (EOFs) of the sample  . (Please note, that
. (Please note, that  is the eigenvector of
is the eigenvector of 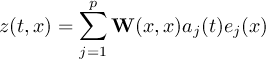 and not the weighted
eigen-vector which would be
and not the weighted
eigen-vector which would be 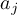 .) Let the corresponding eigenvalues be denoted
.) Let the corresponding eigenvalues be denoted  . The vectors
. The vectors  are
spatial patterns which explain a certain amount of variance of the time series
are
spatial patterns which explain a certain amount of variance of the time series 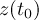 that is related linearly to
that is related linearly to
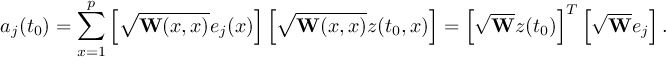 . Thus, the spatial pattern defined by the first eigenvector (the one with the largest eigenvalue ) is the pattern
which explains a maximum possible amount of variance of the sample
. Thus, the spatial pattern defined by the first eigenvector (the one with the largest eigenvalue ) is the pattern
which explains a maximum possible amount of variance of the sample 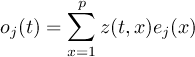 . The orthonormality of eigenvectors
reads as
. The orthonormality of eigenvectors
reads as
![∑p [∘ ------- ] [∘------- ] ∑p { 0 if j ⁄= k
W (x,x)ej(x) W (x,x)ek(x) = W (x,x)ej(x)ek(x) =
x=1 x=1 1 if j = k](cdo163x.png)
If all EOFs 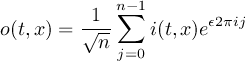 with
with  are calculated, the data can be reconstructed from
are calculated, the data can be reconstructed from
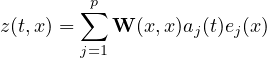
where 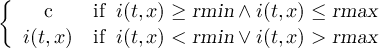 are called the principal components or principal coefficients or EOF coefficients of
are called the principal components or principal coefficients or EOF coefficients of 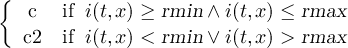 . These
coefficients - as readily seen from above - are calculated as the projection of an EOF
. These
coefficients - as readily seen from above - are calculated as the projection of an EOF  onto a time step of the
data sample
onto a time step of the
data sample 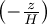 as
as
![∑p [∘ ------- ][∘ ------- ] [√ --- ]T [√ --- ]
aj(t0) = W (x,x)ej(x) W (x,x)z(t0,x) = Wz (t0) Wej .
x=1](cdo171x.png)
Here is a short overview of all operators in this section:
| eof | Calculate EOFs in spatial or time space |
| eoftime | Calculate EOFs in time space |
| eofspatial | Calculate EOFs in spatial space |
| eof3d | Calculate 3-Dimensional EOFs in time space |
| eofcoeff | Calculate principal coefficients of EOFs |
2.11.1 EOF - Empirical Orthogonal Functions
Synopsis
<operator>,neof infile outfile1 outfile2
Description
This module calculates empirical orthogonal functions of the data in infile as the eigen values of the scatter matrix (covariance matrix) S of the data sample z(t). A more detailed description can be found above.
Please note, that the input data are assumed to be anomalies.
If operator eof is chosen, the EOFs are computed in either time or spatial space, whichever is the fastest. If the user already knows, which computation is faster, the module can be forced to perform a computation in time- or gridspace by using the operators eoftime or eofspatial, respectively. This can enhance performance, especially for very long time series, where the number of timesteps is larger than the number of grid-points. Data in infile are assumed to be anomalies. If they are not, the behavior of this module is not well defined. After execution outfile1 will contain all eigen-values and outfile2 the eigenvectors e_j. All EOFs and eigen-values are computed. However, only the first neof EOFs are written to outfile2. Nonetheless, outfile1 contains all eigen-values.
Missing values are not fully supported. Support is only checked for non-changing masks of missing values in time. Although there still will be results, they are not trustworthy, and a warning will occur. In the latter case we suggest to replace missing values by 0 in infile.
Operators
- eof
-
Calculate EOFs in spatial or time space
- eoftime
-
Calculate EOFs in time space
- eofspatial
-
Calculate EOFs in spatial space
- eof3d
-
Calculate 3-Dimensional EOFs in time space
Parameter
- neof
-
INTEGER Number of eigen functions
Environment
- CDO_SVD_MODE
-
Is used to choose the algorithm for eigenvalue calculation. Options are ’jacobi’ for a one-sided parallel jacobi-algorithm (only executed in parallel if -P flag is set) and ’danielson_lanczos’ for a non-parallel d/l algorithm. The default setting is ’jacobi’.
- CDO_WEIGHT_MODE
-
It is used to set the weight mode. The default is ’off’. Set it to ’on’ for a weighted version.
- MAX_JACOBI_ITER
-
Is the maximum integer number of annihilation sweeps that is executed if the jacobi-algorithm is used to compute the eigen values. The default value is 12.
- FNORM_PRECISION
-
Is the Frobenius norm of the matrix consisting of an annihilation pair of eigenvectors that is used to determine if the eigenvectors have reached a sufficient level of convergence. If all annihilation-pairs of vectors have a norm below this value, the computation is considered to have converged properly. Otherwise, a warning will occur. The default value 1e-12.
Example
To calculate the first 40 EOFs of a data-set containing anomalies use:
cdo eof,40 infile outfile1 outfile2
If the dataset does not containt anomalies, process them first, and use:
cdo sub infile1 -timmean infile1 anom_file cdo eof,40 anom_file outfile1 outfile2
2.11.2 EOFCOEFF - Principal coefficients of EOFs
Synopsis
eofcoeff infile1 infile2 obase
Description
This module calculates the time series of the principal coefficients for given EOF (empirical orthogonal functions) and data. Time steps in infile1 are assumed to be the EOFs, time steps in infile2 are assumed to be the time series. Note, that this operator calculates a non weighted dot product of the fields in infile1 and infile2. For consistency set the environment variable CDO_WEIGHT_MODE=off when using eof or eof3d. Given a set of EOFs e_j and a time series of data z(t) with p entries for each timestep from which e_j have been calculated, this operator calculates the time series of the projections of data onto each EOF
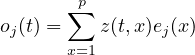
There will be a seperate file o_j for the principal coefficients of each EOF.
As the EOFs e_j are uncorrelated, so are their principal coefficients, i.e.
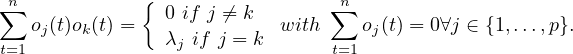
There will be a separate file containing a time series of principal coefficients with time information from infile2 for each EOF in infile1. Output files will be numbered as <obase><neof><suffix> where neof+1 is the number of the EOF (timestep) in infile1 and suffix is the filename extension derived from the file format.
Environment
- CDO_FILE_SUFFIX
-
Set the default file suffix. This suffix will be added to the output file names instead of the filename extension derived from the file format. Set this variable to NULL to disable the adding of a file suffix.
Example
To calculate principal coefficients of the first 40 EOFs of anom_file, and write them to files beginning with obase, use:
export CDO_WEIGHT_MODE=off cdo eof,40 anom_file eval_file eof_file cdo eofcoeff eof_file anom_file obase
The principal coefficients of the first EOF will be in the file obase000000.nc (and so forth for higher EOFs, nth EOF will be in obase<n-1>).
If the dataset infile does not containt anomalies, process them first, and use:
export CDO_WEIGHT_MODE=off cdo sub infile -timmean infile anom_file cdo eof,40 anom_file eval_file eof_file cdo eofcoeff eof_file anom_file obase
2.12 Interpolation
This section contains modules to interpolate datasets. There are several operators to interpolate horizontal fields to a new grid. Some of those operators can handle only 2D fields on a regular rectangular grid. Vertical interpolation of 3D variables is possible from hybrid model levels to height or pressure levels. Interpolation in time is possible between time steps and years.
Here is a short overview of all operators in this section:
| remapbil | Bilinear interpolation |
| genbil | Generate bilinear interpolation weights |
| remapbic | Bicubic interpolation |
| genbic | Generate bicubic interpolation weights |
| remapnn | Nearest neighbor remapping |
| gennn | Generate nearest neighbor remap weights |
| remapdis | Distance weighted average remapping |
| gendis | Generate distance weighted average remap weights |
| remapcon | First order conservative remapping |
| gencon | Generate 1st order conservative remap weights |
| remaplaf | Largest area fraction remapping |
| genlaf | Generate largest area fraction remap weights |
| remap | Grid remapping |
| remapeta | Remap vertical hybrid level |
| ml2pl | Model to pressure level interpolation |
| ml2hl | Model to height level interpolation |
| ap2pl | Air pressure to pressure level interpolation |
| gh2hl | Geometric height to height level interpolation |
| intlevel | Linear level interpolation |
| intlevel3d | Linear level interpolation onto a 3D vertical coordinate |
| intlevelx3d | like intlevel3d but with extrapolation |
| inttime | Interpolation between timesteps |
| intntime | Interpolation between timesteps |
| intyear | Interpolation between two years |
2.12.1 REMAPBIL - Bilinear interpolation
Synopsis
remapbil,grid infile outfile
genbil,grid[,map3d] infile outfile
Description
This module contains operators for a bilinear remapping of fields between grids in spherical coordinates. The interpolation is based on an adapted SCRIP library version. For a detailed description of the interpolation method see [SCRIP]. This interpolation method only works on quadrilateral curvilinear source grids. Below is a schematic illustration of the bilinear remapping:
The figure on the left side shows the input data on a regular lon/lat source grid and on the right side the remapped result on an unstructured triangular target grid. The figure in the middle shows the input data with the target grid. Grid cells with missing value are grey colored.
Operators
- remapbil
-
Bilinear interpolation
Performs a bilinear interpolation on all input fields. - genbil
-
Generate bilinear interpolation weights
Generates bilinear interpolation weights for the first input field and writes the result to a file. The format of this file is NetCDF following the SCRIP convention. Use the operator remap to apply this remapping weights to a data file with the same source grid. Set the parameter map3d=true to generate all mapfiles of the first 3D field with varying masks. In this case the mapfiles will be named <outfile><xxx>.nc. xxx will have five digits with the number of the mapfile.
Parameter
- grid
-
STRING Target grid description file or name
- map3d
-
BOOL Generate all mapfiles of the first 3D field
Environment
- REMAP_EXTRAPOLATE
-
This variable is used to switch the extrapolation feature ’on’ or ’off’. By default the extrapolation is enabled for circular grids.
Example
Say infile contains fields on a quadrilateral curvilinear grid. To remap all fields bilinear to a regular Gaussian F32 grid, type:
cdo remapbil,F32 infile outfile
2.12.2 REMAPBIC - Bicubic interpolation
Synopsis
remapbic,grid infile outfile
genbic,grid[,map3d] infile outfile
Description
This module contains operators for a bicubic remapping of fields between grids in spherical coordinates. The interpolation is based on an adapted SCRIP library version. For a detailed description of the interpolation method see [SCRIP]. This interpolation method only works on quadrilateral curvilinear source grids. Below is a schematic illustration of the bicubic remapping:
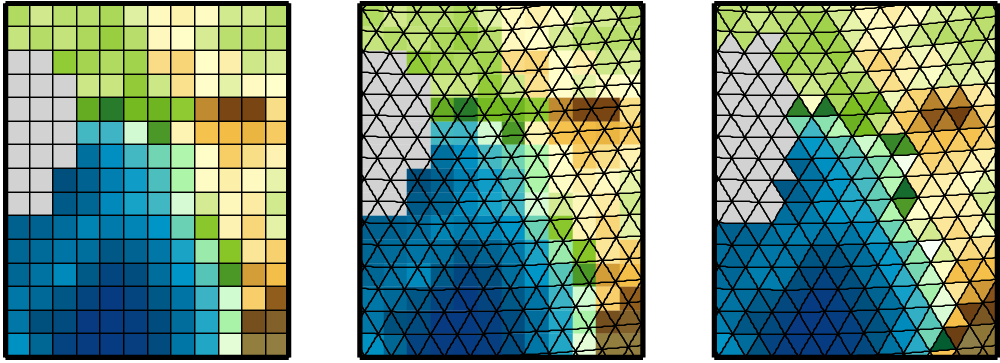
The figure on the left side shows the input data on a regular lon/lat source grid and on the right side the remapped result on an unstructured triangular target grid. The figure in the middle shows the input data with the target grid. Grid cells with missing value are grey colored.
Operators
- remapbic
-
Bicubic interpolation
Performs a bicubic interpolation on all input fields. - genbic
-
Generate bicubic interpolation weights
Generates bicubic interpolation weights for the first input field and writes the result to a file. The format of this file is NetCDF following the SCRIP convention. Use the operator remap to apply this remapping weights to a data file with the same source grid. Set the parameter map3d=true to generate all mapfiles of the first 3D field with varying masks. In this case the mapfiles will be named <outfile><xxx>.nc. xxx will have five digits with the number of the mapfile.
Parameter
- grid
-
STRING Target grid description file or name
- map3d
-
BOOL Generate all mapfiles of the first 3D field
Environment
- REMAP_EXTRAPOLATE
-
This variable is used to switch the extrapolation feature ’on’ or ’off’. By default the extrapolation is enabled for circular grids.
Example
Say infile contains fields on a quadrilateral curvilinear grid. To remap all fields bicubic to a regular Gaussian F32 grid, type:
cdo remapbic,F32 infile outfile
2.12.3 REMAPNN - Nearest neighbor remapping
Synopsis
remapnn,grid infile outfile
gennn,grid[,map3d] infile outfile
Description
This module contains operators for a nearest neighbor remapping of fields between grids in spherical coordinates. Below is a schematic illustration of the nearest neighbor remapping:
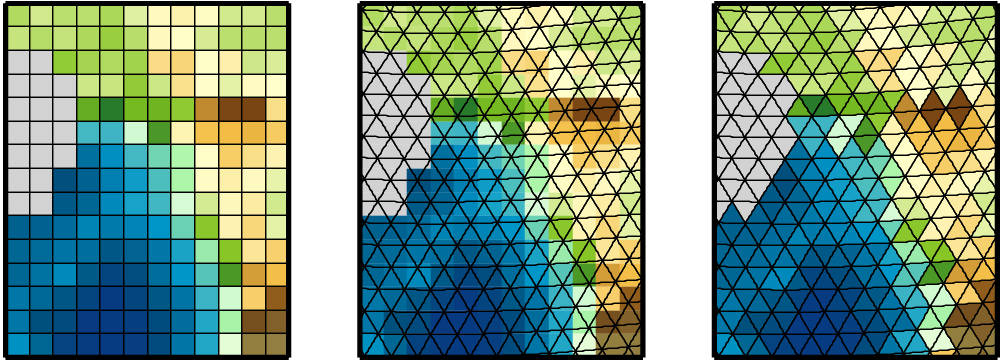
The figure on the left side shows the input data on a regular lon/lat source grid and on the right side the remapped result on an unstructured triangular target grid. The figure in the middle shows the input data with the target grid. Grid cells with missing value are grey colored.
Operators
- remapnn
-
Nearest neighbor remapping
Performs a nearest neighbor remapping on all input fields. - gennn
-
Generate nearest neighbor remap weights
Generates nearest neighbor remapping weights for the first input field and writes the result to a file. The format of this file is NetCDF following the SCRIP convention. Use the operator remap to apply this remapping weights to a data file with the same source grid. Set the parameter map3d=true to generate all mapfiles of the first 3D field with varying masks. In this case the mapfiles will be named <outfile><xxx>.nc. xxx will have five digits with the number of the mapfile.
Parameter
- grid
-
STRING Target grid description file or name
- map3d
-
BOOL Generate all mapfiles of the first 3D field
Environment
- REMAP_EXTRAPOLATE
-
This variable is used to switch the extrapolation feature ’on’ or ’off’. By default the extrapolation is enabled for this remapping method.
- CDO_GRIDSEARCH_RADIUS
-
Grid search radius in degree, default 180 degree.
2.12.4 REMAPDIS - Distance weighted average remapping
Synopsis
remapdis,grid[,neighbors] infile outfile
gendis,grid[,neighbors[,map3d]] infile outfile
Description
This module contains operators for an inverse distance weighted average remapping of the four nearest neighbor values of fields between grids in spherical coordinates. The default number of 4 neighbors can be changed with the neighbors parameter. Below is a schematic illustration of the distance weighted average remapping:
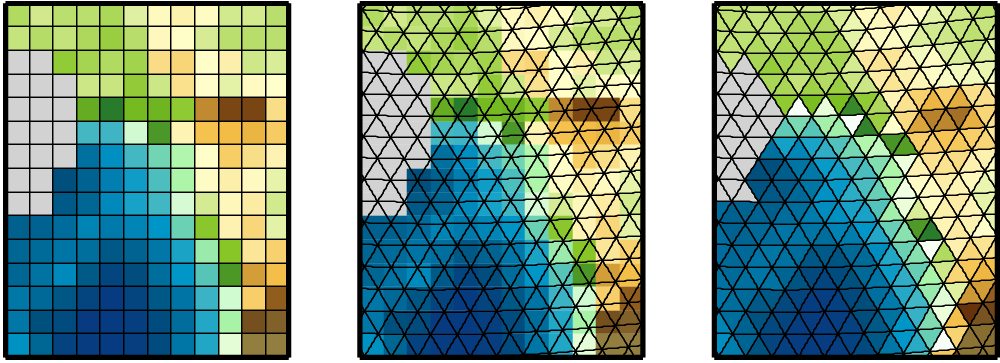
The figure on the left side shows the input data on a regular lon/lat source grid and on the right side the remapped result on an unstructured triangular target grid. The figure in the middle shows the input data with the target grid. Grid cells with missing value are grey colored.
Operators
- remapdis
-
Distance weighted average remapping
Performs an inverse distance weighted averaged remapping of the nearest neighbor values on all input fields. - gendis
-
Generate distance weighted average remap weights
Generates distance weighted averaged remapping weights of the nearest neighbor values for the first input field and writes the result to a file. The format of this file is NetCDF following the SCRIP convention. Use the operator remap to apply this remapping weights to a data file with the same source grid. Set the parameter map3d=true to generate all mapfiles of the first 3D field with varying masks. In this case the mapfiles will be named <outfile><xxx>.nc. xxx will have five digits with the number of the mapfile.
Parameter
- grid
-
STRING Target grid description file or name
- neighbors
-
INTEGER Number of nearest neighbors [default: 4]
- map3d
-
BOOL Generate all mapfiles of the first 3D field
Environment
- REMAP_EXTRAPOLATE
-
This variable is used to switch the extrapolation feature ’on’ or ’off’. By default the extrapolation is enabled for this remapping method.
- CDO_GRIDSEARCH_RADIUS
-
Grid search radius in degree, default 180 degree.
2.12.5 REMAPCON - First order conservative remapping
Synopsis
remapcon,grid infile outfile
gencon,grid[,map3d] infile outfile
Description
This module contains operators for a first order conservative remapping of fields between grids in spherical coordinates. The operators in this module uses code from the YAC software package to compute the conservative remapping weights. For a detailed description of the interpolation method see [YAC]. The interpolation method is completely general and can be used for any grid on a sphere. The search algorithm for the conservative remapping requires that no grid cell occurs more than once. Below is a schematic illustration of the 1st order conservative remapping:
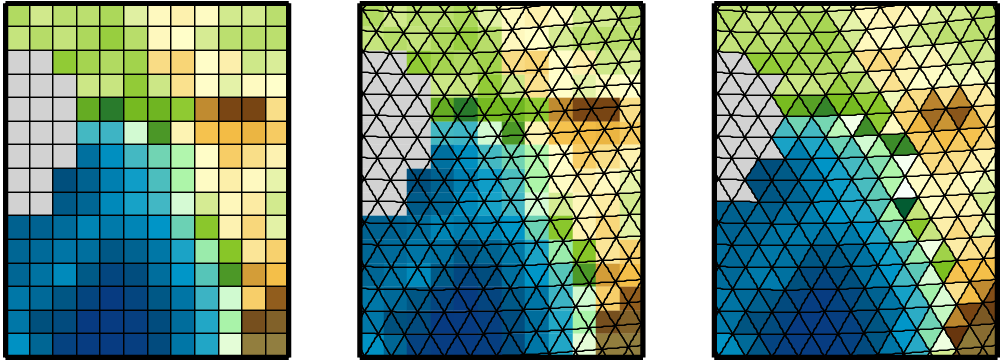
The figure on the left side shows the input data on a regular lon/lat source grid and on the right side the remapped result on an unstructured triangular target grid. The figure in the middle shows the input data with the target grid. Grid cells with missing value are grey colored.
Operators
- remapcon
-
First order conservative remapping
Performs a first order conservative remapping on all input fields. - gencon
-
Generate 1st order conservative remap weights
Generates first order conservative remapping weights for the first input field and writes the result to a file. The format of this file is NetCDF following the SCRIP convention. Use the operator remap to apply this remapping weights to a data file with the same source grid. Set the parameter map3d=true to generate all mapfiles of the first 3D field with varying masks. In this case the mapfiles will be named <outfile><xxx>.nc. xxx will have five digits with the number of the mapfile.
Parameter
- grid
-
STRING Target grid description file or name
- map3d
-
BOOL Generate all mapfiles of the first 3D field
Environment
- CDO_REMAP_NORM
-
This variable is used to choose the normalization of the conservative interpolation. By default CDO_REMAP_NORM is set to ’fracarea’. ’fracarea’ uses the sum of the non-masked source cell intersected areas to normalize each target cell field value. This results in a reasonable flux value but the flux is not locally conserved. The option ’destarea’ uses the total target cell area to normalize each target cell field value. Local flux conservation is ensured, but unreasonable flux values may result.
- REMAP_AREA_MIN
-
This variable is used to set the minimum destination area fraction. The default of this variable is 0.0.
Example
Say infile contains fields on a quadrilateral curvilinear grid. To remap all fields conservative to a regular Gaussian F32 grid, type:
cdo remapcon,F32 infile outfile
2.12.6 REMAPLAF - Largest area fraction remapping
Synopsis
<operator>,grid infile outfile
Description
This module contains operators for a largest area fraction remapping of fields between grids in spherical coordinates. The operators in this module uses code from the YAC software package to compute the largest area fraction. For a detailed description of the interpolation method see [YAC]. The interpolation method is completely general and can be used for any grid on a sphere. The search algorithm for this remapping method requires that no grid cell occurs more than once. Below is a schematic illustration of the largest area fraction conservative remapping:
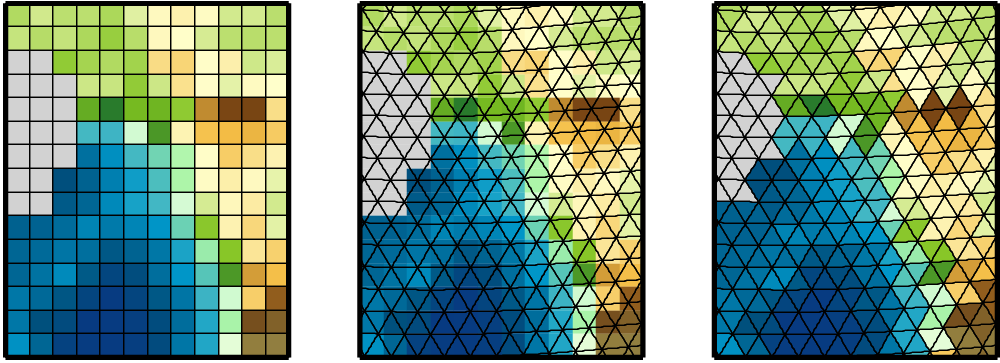
The figure on the left side shows the input data on a regular lon/lat source grid and on the right side the remapped result on an unstructured triangular target grid. The figure in the middle shows the input data with the target grid. Grid cells with missing value are grey colored.
Operators
- remaplaf
-
Largest area fraction remapping
Performs a largest area fraction remapping on all input fields. - genlaf
-
Generate largest area fraction remap weights
Generates largest area fraction remapping weights for the first input field and writes the result to a file. The format of this file is NetCDF following the SCRIP convention. Use the operator remap to apply this remapping weights to a data file with the same source grid.
Parameter
- grid
-
STRING Target grid description file or name
Environment
- REMAP_AREA_MIN
-
This variable is used to set the minimum destination area fraction. The default of this variable is 0.0.
2.12.7 REMAP - Grid remapping
Synopsis
remap,grid,weights infile outfile
Description
Interpolation between different horizontal grids can be a very time-consuming process. Especially if the data are on an unstructured and/or a large grid. In this case the interpolation process can be split into two parts. Firstly the generation of the interpolation weights, which is the most time-consuming part. These interpolation weights can be reused for every remapping process with the operator remap. This operator remaps all input fields to a new horizontal grid. The remap type and the interpolation weights of one input grid are read from a NetCDF file. More weights are computed if the input fields are on different grids. The NetCDF file with the weights should follow the [SCRIP] convention. Normally these weights come from a previous call to one of the genXXX operators (e.g. genbil) or were created by the original SCRIP package.
Parameter
- grid
-
STRING Target grid description file or name
- weights
-
STRING Interpolation weights (SCRIP NetCDF file)
Environment
- CDO_REMAP_NORM
-
This variable is used to choose the normalization of the conservative interpolation. By default CDO_REMAP_NORM is set to ’fracarea’. ’fracarea’ uses the sum of the non-masked source cell intersected areas to normalize each target cell field value. This results in a reasonable flux value but the flux is not locally conserved. The option ’destarea’ uses the total target cell area to normalize each target cell field value. Local flux conservation is ensured, but unreasonable flux values may result.
- REMAP_EXTRAPOLATE
-
This variable is used to switch the extrapolation feature ’on’ or ’off’. By default the extrapolation is enabled for remapdis, remapnn and for circular grids.
- REMAP_AREA_MIN
-
This variable is used to set the minimum destination area fraction. The default of this variable is 0.0.
- CDO_GRIDSEARCH_RADIUS
-
Grid search radius in degree, default 180 degree.
Example
Say infile contains fields on a quadrilateral curvilinear grid. To remap all fields bilinear to a regular Gaussian F32 grid use:
cdo genbil,F32 infile remapweights.nc cdo remap,F32,remapweights.nc infile outfile
The result will be the same as:
cdo remapbil,F32 infile outfile
2.12.8 REMAPETA - Remap vertical hybrid level
Synopsis
remapeta,vct[,oro] infile outfile
Description
This operator interpolates between different vertical hybrid levels. This include the preparation of consistent data for the free atmosphere. The procedure for the vertical interpolation is based on the HIRLAM scheme and was adapted from [INTERA]. The vertical interpolation is based on the vertical integration of the hydrostatic equation with few adjustments. The basic tasks are the following one:
-
at first integration of hydrostatic equation
-
extrapolation of surface pressure
-
Planetary Boundary-Layer (PBL) proutfile interpolation
-
interpolation in free atmosphere
-
merging of both proutfiles
-
final surface pressure correction
The vertical interpolation corrects the surface pressure. This is simply a cut-off or an addition of air mass. This mass correction should not influence the geostrophic velocity field in the middle troposhere. Therefore the total mass above a given reference level is conserved. As reference level the geopotential height of the 400 hPa level is used. Near the surface the correction can affect the vertical structure of the PBL. Therefore the interpolation is done using the potential temperature. But in the free atmosphere above a certain n (n=0.8 defining the top of the PBL) the interpolation is done linearly. After the interpolation both proutfiles are merged. With the resulting temperature/pressure correction the hydrostatic equation is integrated again and adjusted to the reference level finding the final surface pressure correction. A more detailed description of the interpolation can be found in [INTERA]. This operator requires all variables on the same horizontal grid.
Parameter
- vct
-
STRING File name of an ASCII dataset with the vertical coordinate table
- oro
-
STRING File name with the orography (surf. geopotential) of the target dataset (optional)
Environment
- REMAPETA_PTOP
-
Sets the minimum pressure level for condensation. Above this level the humidity is set to the constant 1.E-6. The default value is 0 Pa.
Note
The code numbers or the variable names of the required parameter have to follow the [ECHAM] convention.
Use the sinfo command to test if your vertical coordinate system is recognized as hybrid system.
In case remapeta complains about not finding any data on hybrid model levels you may wish to use the setzaxis command to generate a zaxis description which conforms to the ECHAM convention. See section "1.4 Z-axis description" for an example how to define a hybrid Z-axis.
Example
To remap between different hybrid model level data use:
cdo remapeta,vct infile outfile
Here is an example vct file with 19 hybrid model level:
0 0.00000000000000000 0.00000000000000000 1 2000.00000000000000000 0.00000000000000000 2 4000.00000000000000000 0.00000000000000000 3 6046.10937500000000000 0.00033899326808751 4 8267.92968750000000000 0.00335718691349030 5 10609.51171875000000000 0.01307003945112228 6 12851.10156250000000000 0.03407714888453484 7 14698.50000000000000000 0.07064980268478394 8 15861.12890625000000000 0.12591671943664551 9 16116.23828125000000000 0.20119541883468628 10 15356.92187500000000000 0.29551959037780762 11 13621.46093750000000000 0.40540921688079834 12 11101.55859375000000000 0.52493220567703247 13 8127.14453125000000000 0.64610791206359863 14 5125.14062500000000000 0.75969839096069336 15 2549.96899414062500000 0.85643762350082397 16 783.19506835937500000 0.92874687910079956 17 0.00000000000000000 0.97298520803451538 18 0.00000000000000000 0.99228149652481079 19 0.00000000000000000 1.00000000000000000
2.12.9 VERTINTML - Vertical interpolation
Synopsis
ml2pl,plevels infile outfile
ml2hl,hlevels infile outfile
Description
Interpolates 3D variables on hybrid sigma pressure level to pressure or height levels. A basic linear method is used for interpolation. To calculate the pressure on model levels, the a and b coefficients defining the model levels and the surface pressure are required. The a and b coefficients are normally part of the model level data. If not available, the surface pressure can be derived from the logarithm of the surface pressure. To extrapolate the temperature, the surface geopotential is also needed. The geopotential height must be present at the hybrid layer interfaces (model half-layers)! All needed variables are identified by their GRIB1 code number or NetCDF CF standard name. Supported parameter tables are: WMO standard table number 2 and ECMWF local table number 128.
| Name | Units | GRIB1 code | CF standard name |
| log surface pressure | Pa | 152 | |
| surface pressure | Pa | 134 | surface_air_pressure |
| air temperature | K | 130 | air_temperature |
| surface geopotential | m2 s-2 | 129 | surface_geopotential |
| geopotential height | m | 156 | geopotential_height |
Use the alias ml2plx/ml2hlx to fill in missing values with the next available value of the same vertical column. Only the temperature is extrapolated in this case. The extrapolation method originates from the ECHAM postprocessing. This operator requires all variables on the same horizontal grid. Missing values in the input data are not supported.
Operators
- ml2pl
-
Model to pressure level interpolation
Interpolates 3D variables on hybrid sigma pressure level to pressure level. - ml2hl
-
Model to height level interpolation
Interpolates 3D variables on hybrid sigma pressure level to height level. The procedure is the same as for the operator ml2pl except for the pressure levels being calculated from the heights by: plevel = 101325 * exp(hlevel∕ - 7000)
Parameter
- plevels
-
FLOAT Pressure levels in pascal
- hlevels
-
FLOAT Height levels in meter
Note
This is a specific implementation for data from the ECHAM model, it may not work with data from other sources. The components of the hybrid coordinate must always be avaiable at the hybrid layer interfaces even if the data is defined at the hybrid layer midpoints.
Example
To interpolate hybrid model level data to pressure levels of 925, 850, 500 and 200 hPa use:
cdo ml2pl,92500,85000,50000,20000 infile outfile
2.12.10 VERTINTAP - Vertical pressure interpolation
Synopsis
ap2pl,levels infile outfile
Description
Interpolate 3D variables on hybrid sigma height coordinates to pressure levels. A basic linear method is used for interpolation. The input file must contain the 3D air pressure in pascal. The air pressure is identified by the NetCDF CF standard name air_pressure. Use the alias ap2plx to fill in missing values with the next available value of the same vertical column. This operator requires all variables on the same horizontal grid. Missing values in the input data are not supported.
Parameter
- levels
-
FLOAT Comma-separated list of pressure levels in pascal
Note
This is a specific implementation for NetCDF files from the ICON model, it may not work with data from other sources.
Example
To interpolate 3D variables on hybrid sigma height level to pressure levels of 925, 850, 500 and 200 hPa use:
cdo ap2pl,92500,85000,50000,20000 infile outfile
2.12.11 VERTINTGH - Vertical height interpolation
Synopsis
gh2hl,levels infile outfile
Description
Interpolate 3D variables on hybrid sigma height coordinates to height levels. A basic linear method is used for interpolation. The input file must contain the 3D geometric height in meter. The geometric height is identified by the NetCDF CF standard name geometric_height_at_full_level_center. Use the alias gh2hlx to fill in missing values with the next available value of the same vertical column. This operator requires all variables on the same horizontal grid. Missing values in the input data are not supported.
Parameter
- levels
-
FLOAT Comma-separated list of height levels in meter
Note
This is a specific implementation for NetCDF files from the ICON model, it may not work with data from other sources.
Example
To interpolate 3D variables on hybrid sigma height level to height levels of 20, 100, 500, 1000, 5000, 10000 and 20000 meter use:
cdo gh2hl,20,100,500,1000,5000,10000,20000 infile outfile
2.12.12 INTLEVEL - Linear level interpolation
Synopsis
intlevel,parameter infile outfile
Description
This operator performs a linear vertical interpolation of 3D variables. The 1D target levels can be specified with the level parameter or read in via a Z-axis description file.
Parameter
- level
-
FLOAT Comma-separated list of target levels
- zdescription
-
STRING Path to a file containing a description of the Z-axis
- zvarname
-
STRING Use zvarname as the vertical 3D source coordinate instead of the 1D coordinate variable
- extrapolate
-
BOOL Fill target layers out of the source layer range with the nearest source layer
Example
To interpolate 3D variables on height levels to a new set of height levels use:
cdo intlevel,level=10,50,100,500,1000 infile outfile
2.12.13 INTLEVEL3D - Linear level interpolation from/to 3D vertical coordinates
Synopsis
<operator>,tgtcoordinate infile1 infile2 outfile
Description
This operator performs a linear vertical interpolation of 3D variables fields with given 3D vertical coordinates. infile1 contains the 3D data variables and infile2 the 3D vertical source coordinate. The parameter tgtcoordinate is a datafile with the 3D vertical target coordinate.
Operators
- intlevel3d
-
Linear level interpolation onto a 3D vertical coordinate
- intlevelx3d
-
like intlevel3d but with extrapolation
Parameter
- tgtcoordinate
-
STRING filename for 3D vertical target coordinates
Example
To interpolate 3D variables from one set of 3D height levels into another one where
-
infile2 contains a single 3D variable, which represents the source 3D vertical coordinate
-
infile1 contains the source data, which the vertical coordinate from infile2 belongs to
-
tgtcoordinate only contains the target 3D height levels
cdo intlevel3d,tgtcoordinate infile1 infile2 outfile
2.12.14 INTTIME - Time interpolation
Synopsis
inttime,date,time[,inc] infile outfile
intntime,n infile outfile
Description
This module performs linear interpolation between timesteps. Interpolation is only performed if both values exist. If both values are missing values, the result is also a missing value. If only one value exists, it is taken if the time weighting is greater than or equal to 0.5. So no new value will be created at existing time steps, if the value is missing there.
Operators
- inttime
-
Interpolation between timesteps
This operator creates a new dataset by linear interpolation between timesteps. The user has to define the start date/time with an optional increment. - intntime
-
Interpolation between timesteps
This operator performs linear interpolation between timesteps. The user has to define the number of timesteps from one timestep to the next.
Parameter
- date
-
STRING Start date (format YYYY-MM-DD)
- time
-
STRING Start time (format hh:mm:ss)
- inc
-
STRING Optional increment (seconds, minutes, hours, days, months, years) [default: 0hour]
- n
-
INTEGER Number of timesteps from one timestep to the next
Example
Assumed a 6 hourly dataset starts at 1987-01-01 12:00:00. To interpolate this time series to a one hourly dataset use:
cdo inttime,1987-01-01,12:00:00,1hour infile outfile
2.12.15 INTYEAR - Year interpolation
Synopsis
intyear,years infile1 infile2 obase
Description
This operator performs linear interpolation between two years, timestep by timestep. The input files need to have the same structure with the same variables. The output files will be named <obase><yyyy><suffix> where yyyy will be the year and suffix is the filename extension derived from the file format.
Parameter
- years
-
INTEGER Comma-separated list or first/last[/inc] range of years
Environment
- CDO_FILE_SUFFIX
-
Set the default file suffix. This suffix will be added to the output file names instead of the filename extension derived from the file format. Set this variable to NULL to disable the adding of a file suffix.
Note
This operator needs to open all output files simultaneously. The maximum number of open files depends on the operating system!
Example
Assume there are two monthly mean datasets over a year. The first dataset has 12 timesteps for the year 1985 and the second one for the year 1990. To interpolate the years between 1985 and 1990 month by month use:
cdo intyear,1986,1987,1988,1989 infile1 infile2 year
Example result of ’dir year*’ for NetCDF datasets:
year1986.nc year1987.nc year1988.nc year1989.nc
2.13 Transformation
This section contains modules to perform spectral transformations.
Here is a short overview of all operators in this section:
| sp2gp | Spectral to gridpoint |
| gp2sp | Gridpoint to spectral |
| sp2sp | Spectral to spectral |
| dv2ps | D and V to velocity potential and stream function |
| dv2uv | Divergence and vorticity to U and V wind |
| uv2dv | U and V wind to divergence and vorticity |
| fourier | Fourier transformation |
2.13.1 SPECTRAL - Spectral transformation
Synopsis
<operator>[,type|trunc] infile outfile
Description
This module transforms fields on a global regular Gaussian grid to spectral coefficients and vice versa. The transformation is achieved by applying Fast Fourier Transformation (FFT) first and direct Legendre Transformation afterwards in gp2sp. In sp2gp the inverse Legendre Transformation and inverse FFT are used. Missing values are not supported.
The relationship between the spectral resolution, governed by the truncation number T, and the grid resolution depends on the number of grid points at which the shortest wavelength field is represented. For a grid with 2N points between the poles (so 4N grid points in total around the globe) the relationship is:
linear grid: the shortest wavelength is represented by 2 grid points -> 4N ≃ 2(TL + 1)
quadratic grid: the shortest wavelength is represented by 3 grid points -> 4N ≃ 3(TQ + 1)
cubic grid: the shortest wavelength is represented by 4 grid points -> 4N ≃ 4(TC + 1)
The quadratic grid is used by ECHAM and ERA15. ERA40 is using a linear Gaussian grid reflected by the TL notation.
The following table shows the calculation of the number of latitudes and the triangular truncation for the different grid types:
| Gridtype | Number of latitudes: nlat | Triangular truncation: ntr |
| linear | NINT((ntr*2 + 1)/2) | (nlat*2 - 1) / 2 |
| quadratic | NINT((ntr*3 + 1)/2) | (nlat*2 - 1) / 3 |
| cubic | NINT((ntr*4 + 1)/2) | (nlat*2 - 1) / 4 |
Operators
- sp2gp
-
Spectral to gridpoint
Convert all spectral fields to a global regular Gaussian grid. The optional parameter trunc must be greater than the input truncation. - gp2sp
-
Gridpoint to spectral
Convert all Gaussian gridpoint fields to spectral fields. The optional parameter trunc must be lower than the input truncation.
Parameter
- type
-
STRING Type of the grid: quadratic, linear, cubic (default: type=quadratic)
- trunc
-
STRING Triangular truncation
Note
To speed up the calculations, the Legendre polynoms are kept in memory. This requires a relatively large amount of memory. This is for example 12GB for T1279 data.
Example
To transform spectral coefficients from T106 to F80 regular Gaussian grid use:
cdo sp2gp infile outfile
To transform spectral coefficients from TL159 to F80 regular Gaussian grid use:
cdo sp2gp,type=linear infile outfile
2.13.2 SPECCONV - Spectral conversion
Synopsis
sp2sp,trunc infile outfile
Description
Changed the triangular truncation of all spectral fields. This operator performs downward conversion by cutting the resolution. Upward conversions are achieved by filling in zeros.
Parameter
- trunc
-
INTEGER New spectral resolution
2.13.3 WIND2 - D and V to velocity potential and stream function
Synopsis
dv2ps infile outfile
Description
Calculate spherical harmonic coefficients of velocity potential and stream function from spherical harmonic coefficients of relative divergence and vorticity. The divergence and vorticity need to have the names sd and svo or code numbers 155 and 138.
2.13.4 WIND - Wind transformation
Synopsis
<operator>[,gridtype] infile outfile
Description
This module converts relative divergence and vorticity to U and V wind and vice versa. Divergence and vorticity are spherical harmonic coefficients in spectral space and U and V are on a global regular Gaussian grid. The Gaussian latitudes need to be ordered from north to south. Missing values are not supported.
The relationship between the spectral resolution, governed by the truncation number T, and the grid resolution depends on the number of grid points at which the shortest wavelength field is represented. For a grid with 2N points between the poles (so 4N grid points in total around the globe) the relationship is:
linear grid: the shortest wavelength is represented by 2 grid points -> 4N ≃ 2(TL + 1)
quadratic grid: the shortest wavelength is represented by 3 grid points -> 4N ≃ 3(TQ + 1)
cubic grid: the shortest wavelength is represented by 4 grid points -> 4N ≃ 4(TC + 1)
The quadratic grid is used by ECHAM and ERA15. ERA40 is using a linear Gaussian grid reflected by the TL notation.
The following table shows the calculation of the number of latitudes and the triangular truncation for the different grid types:
| Gridtype | Number of latitudes: nlat | Triangular truncation: ntr |
| linear | NINT((ntr*2 + 1)/2) | (nlat*2 - 1) / 2 |
| quadratic | NINT((ntr*3 + 1)/2) | (nlat*2 - 1) / 3 |
| cubic | NINT((ntr*4 + 1)/2) | (nlat*2 - 1) / 4 |
Operators
- dv2uv
-
Divergence and vorticity to U and V wind
Calculate U and V wind on a Gaussian grid from spherical harmonic coefficients of relative divergence and vorticity. The divergence and vorticity need to have the names sd and svo or code numbers 155 and 138. - uv2dv
-
U and V wind to divergence and vorticity
Calculate spherical harmonic coefficients of relative divergence and vorticity from U and V wind. The U and V wind need to be on a Gaussian grid and need to have the names u and v or the code numbers 131 and 132.
Parameter
- gridtype
-
STRING Type of the grid: quadratic, linear, cubic (default: quadratic)
Note
To speed up the calculations, the Legendre polynoms are kept in memory. This requires a relatively large amount of memory. This is for example 12GB for T1279 data.
Example
Assume a dataset has at least spherical harmonic coefficients of divergence and vorticity. To transform the spectral divergence and vorticity to U and V wind on a Gaussian grid use:
cdo dv2uv infile outfile
2.13.5 FOURIER - Fourier transformation
Synopsis
fourier,epsilon infile outfile
Description
The fourier operator performs the fourier transformation or the inverse fourier transformation of all input fields. If the number of timesteps is a power of 2 then the algorithm of the Fast Fourier Transformation (FFT) is used.
It is
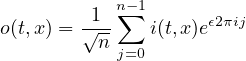
where a user given epsilon = -1 leads to the forward transformation and a user given epsilon = 1 leads to the backward transformation.
If the input stream infile consists only of complex fields, then the fields of outfile, computed by
cdo -f ext fourier,1 -fourier,-1 infile outfile
are the same than that of infile. For real input files see function retocomplex.
Parameter
- epsilon
-
INTEGER -1: forward transformation; 1: backward transformation
Note
Complex numbers can only be stored in NetCDF4 and EXTRA format.
2.14 Import/Export
This section contains modules to import and export data files which can not read or write directly with CDO.
Here is a short overview of all operators in this section:
| import_binary | Import binary data sets |
| import_cmsaf | Import CM-SAF HDF5 files |
| input | ASCII input |
| inputsrv | SERVICE ASCII input |
| inputext | EXTRA ASCII input |
| output | ASCII output |
| outputf | Formatted output |
| outputint | Integer output |
| outputsrv | SERVICE ASCII output |
| outputext | EXTRA ASCII output |
| outputtab | Table output |
| gmtxyz | GMT xyz format |
| gmtcells | GMT multiple segment format |
2.14.1 IMPORTBINARY - Import binary data sets
Synopsis
import_binary infile outfile
Description
This operator imports gridded binary data sets via a GrADS data descriptor file. The GrADS data descriptor file contains a complete description of the binary data as well as instructions on where to find the data and how to read it. The descriptor file is an ASCII file that can be created easily with a text editor. The general contents of a gridded data descriptor file are as follows:
-
Filename for the binary data
-
Missing or undefined data value
-
Mapping between grid coordinates and world coordinates
-
Description of variables in the binary data set
A detailed description of the components of a GrADS data descriptor file can be found in [GrADS]. Here is a list of the supported components: BYTESWAPPED, CHSUB, DSET, ENDVARS, FILEHEADER, HEADERBYTES, OPTIONS, TDEF, TITLE, TRAILERBYTES, UNDEF, VARS, XDEF, XYHEADER, YDEF, ZDEF
Note
Only 32-bit IEEE floats are supported for standard binary files!
Example
To convert a binary data file to NetCDF use:
cdo -f nc import_binary infile.ctl outfile.nc
Here is an example of a GrADS data descriptor file:
DSET ^infile.bin OPTIONS sequential UNDEF -9e+33 XDEF 360 LINEAR -179.5 1 YDEF 180 LINEAR -89.5 1 ZDEF 1 LINEAR 1 1 TDEF 1 LINEAR 00:00Z15jun1989 12hr VARS 1 param 1 99 description of the variable ENDVARS
The binary data file infile.bin contains one parameter on a global 1 degree lon/lat grid written with FORTRAN record length headers (sequential).
2.14.2 IMPORTCMSAF - Import CM-SAF HDF5 files
Synopsis
import_cmsaf infile outfile
Description
This operator imports gridded CM-SAF (Satellite Application Facility on Climate Monitoring) HDF5 files. CM-SAF exploits data from polar-orbiting and geostationary satellites in order to provide climate monitoring products of the following parameters:
- Cloud parameters:
-
cloud fraction (CFC), cloud type (CTY), cloud phase (CPH), cloud top height, pressure and temperature (CTH,CTP,CTT), cloud optical thickness (COT), cloud water path (CWP).
- Surface radiation components:
-
Surface albedo (SAL); surface incoming (SIS) and net (SNS) shortwave radiation; surface downward (SDL) and outgoing (SOL) longwave radiation, surface net longwave radiation (SNL) and surface radiation budget (SRB).
- Top-of-atmosphere radiation components:
-
Incoming (TIS) and reflected (TRS) solar radiative flux at top-of-atmosphere. Emitted thermal radiative flux at top-of-atmosphere (TET).
- Water vapour:
-
Vertically integrated water vapour (HTW), layered vertically integrated water vapour and layer mean temperature and relative humidity for 5 layers (HLW), temperature and mixing ratio at 6 pressure levels.
Daily and monthly mean products can be ordered via the CM-SAF web page (www.cmsaf.eu). Products with higher spatial and temporal resolution, i.e. instantaneous swath-based products, are available on request (contact.cmsaf@dwd.de). All products are distributed free-of-charge. More information on the data is available on the CM-SAF homepage (www.cmsaf.eu).
Daily and monthly mean products are provided in equal-area projections. CDO reads the projection parameters from the metadata in the HDF5-headers in order to allow spatial operations like remapping. For spatial operations with instantaneous products on original satellite projection, additional files with arrays of latitudes and longitudes are needed. These can be obtained from CM-SAF together with the data.
Note
To use this operator, it is necessary to build CDO with HDF5 support (version 1.6 or higher). The PROJ library (version 5.0 or higher) is needed for full support of the remapping functionality.
Example
A typical sequence of commands with this operator could look like this:
cdo -f nc remapbil,r360x180 -import_cmsaf cmsaf_product.hdf output.nc
(bilinear remapping to a predefined global grid with 1 deg resolution and conversion to NetCDF).
If you work with CM-SAF data on original satellite project, an additional file with information on geolocation is required, to perform such spatial operations:
cdo -f nc remapbil,r720x360 -setgrid,cmsaf_latlon.h5 -import_cmsaf cmsaf.hdf out.nc
Some CM-SAF data are stored as scaled integer values. For some operations, it could be desirable (or necessary) to increase the accuracy of the converted products:
cdo -b f32 -f nc fldmean -sellonlatbox,0,10,0,10 -remapbil,r720x360 \ -import_cmsaf cmsaf_product.hdf output.nc
2.14.3 INPUT - Formatted input
Synopsis
input,grid[,zaxis] outfile
inputsrv outfile
inputext outfile
Description
This module reads time series of one 2D variable from standard input. All input fields need to have the same horizontal grid. The format of the input depends on the chosen operator.
Operators
- input
-
ASCII input
Reads fields with ASCII numbers from standard input and stores them in outfile. The numbers read are exactly that ones which are written out by the output operator. - inputsrv
-
SERVICE ASCII input
Reads fields with ASCII numbers from standard input and stores them in outfile. Each field should have a header of 8 integers (SERVICE likely). The numbers that are read are exactly that ones which are written out by the outputsrv operator. - inputext
-
EXTRA ASCII input
Read fields with ASCII numbers from standard input and stores them in outfile. Each field should have header of 4 integers (EXTRA likely). The numbers read are exactly that ones which are written out by the outputext operator.
Parameter
- grid
-
STRING Grid description file or name
- zaxis
-
STRING Z-axis description file
Example
Assume an ASCII dataset contains a field on a global regular grid with 32 longitudes and 16 latitudes (512 elements). To create a GRIB1 dataset from the ASCII dataset use:
cdo -f grb input,r32x16 outfile.grb < my_ascii_data
2.14.4 OUTPUT - Formatted output
Synopsis
output infiles
outputf,format[,nelem] infiles
outputint infiles
outputsrv infiles
outputext infiles
Description
This module prints all values of all input datasets to standard output. All input fields need to have the same horizontal grid. All input files need to have the same structure with the same variables. The format of the output depends on the chosen operator.
Operators
- output
-
ASCII output
Prints all values to standard output. Each row has 6 elements with the C-style format "%13.6g". - outputf
-
Formatted output
Prints all values to standard output. The format and number of elements for each row have to be specified by the parameters format and nelem. The default for nelem is 1. - outputint
-
Integer output
Prints all values rounded to the nearest integer to standard output. - outputsrv
-
SERVICE ASCII output
Prints all values to standard output. Each field with a header of 8 integers (SERVICE likely). - outputext
-
EXTRA ASCII output
Prints all values to standard output. Each field with a header of 4 integers (EXTRA likely).
Parameter
- format
-
STRING C-style format for one element (e.g. %13.6g)
- nelem
-
INTEGER Number of elements for each row (default: nelem = 1)
Example
To print all field elements of a dataset formatted with "%8.4g" and 8 values per line use:
cdo outputf,%8.4g,8 infile
Example result of a dataset with one field on 64 grid points:
261.7 262 257.8 252.5 248.8 247.7 246.3 246.1 250.6 252.6 253.9 254.8 252 246.6 249.7 257.9 273.4 266.2 259.8 261.6 257.2 253.4 251 263.7 267.5 267.4 272.2 266.7 259.6 255.2 272.9 277.1 275.3 275.5 276.4 278.4 282 269.6 278.7 279.5 282.3 284.5 280.3 280.3 280 281.5 284.7 283.6 292.9 290.5 293.9 292.6 292.7 292.8 294.1 293.6 293.8 292.6 291.2 292.6 293.2 292.8 291 291.2
2.14.5 OUTPUTTAB - Table output
Synopsis
outputtab,parameter infiles outfile
Description
This operator prints a table of all input datasets to standard output. infiles is an arbitrary number of input files. All input files need to have the same structure with the same variables on different timesteps. All input fields need to have the same horizontal grid.
The contents of the table depends on the chosen parameters. The format of each table parameter is keyname[:len]. len is the optional length of a table entry. The number of significant digits of floating point parameters can be set with the CDO option --precision, the default is 7. Here is a list of all valid keynames:
| Keyname | Type | Description |
| value | FLOAT | Value of the variable [len:8] |
| name | STRING | Name of the variable [len:8] |
| param | STRING | Parameter ID (GRIB1: code[.tabnum]; GRIB2: num[.cat[.dis]]) [len:11] |
| code | INTEGER | Code number [len:4] |
| x | FLOAT | X coordinate of the original grid [len:6] |
| y | FLOAT | Y coordinate of the original grid [len:6] |
| lon | FLOAT | Longitude coordinate in degrees [len:6] |
| lat | FLOAT | Latitude coordinate in degrees [len:6] |
| lev | FLOAT | Vertical level [len:6] |
| xind | INTEGER | Grid x index [len:4] |
| yind | INTEGER | Grid y index [len:4] |
| timestep | INTEGER | Timestep number [len:6] |
| date | STRING | Date (format YYYY-MM-DD) [len:10] |
| time | STRING | Time (format hh:mm:ss) [len:8] |
| year | INTEGER | Year [len:5] |
| month | INTEGER | Month [len:2] |
| day | INTEGER | Day [len:2] |
| nohead | INTEGER | Disable output of header line |
Parameter
- parameter
-
STRING Comma-separated list of keynames, one for each column of the table
Example
To print a table with name, date, lon, lat and value information use:
cdo outputtab,name,date,lon,lat,value infile
Here is an example output of a time series with the yearly mean temperatur at lon=10/lat=53.5:
# name date lon lat value tsurf 1991-12-31 10 53.5 8.83903 tsurf 1992-12-31 10 53.5 8.17439 tsurf 1993-12-31 10 53.5 7.90489 tsurf 1994-12-31 10 53.5 10.0216 tsurf 1995-12-31 10 53.5 9.07798
2.14.6 OUTPUTGMT - GMT output
Synopsis
<operator> infile
Description
This module prints the first field of the input dataset to standard output. The output can be used to generate 2D Lon/Lat plots with [GMT]. The format of the output depends on the chosen operator.
Operators
- gmtxyz
-
GMT xyz format
The operator exports the first field to the GMT xyz ASCII format. The output can be used to create contour plots with the GMT module pscontour. - gmtcells
-
GMT multiple segment format
The operator exports the first field to the GMT multiple segment ASCII format. The output can be used to create shaded gridfill plots with the GMT module psxy.
Example
1) GMT shaded contour plot of a global temperature field with a resolution of 4 degree. The contour interval is 3 with a rainbow color table.
cdo gmtxyz temp > data.gmt makecpt -T213/318/3 -Crainbow > gmt.cpt pscontour -K -JQ0/10i -Rd -I -Cgmt.cpt data.gmt > gmtplot.ps pscoast -O -J -R -Dc -W -B40g20 >> gmtplot.ps
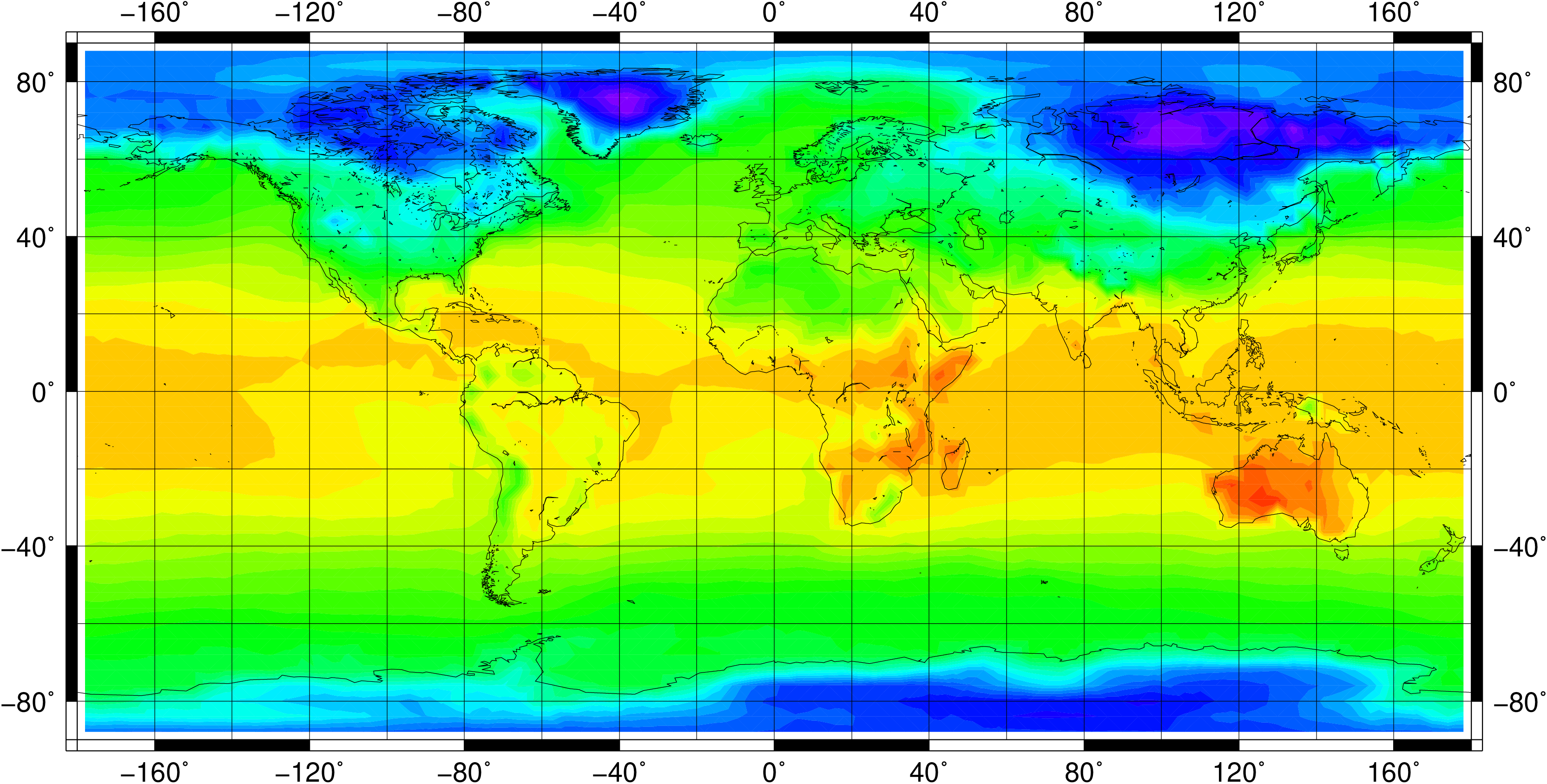
2) GMT shaded gridfill plot of a global temperature field with a resolution of 4 degree. The contour interval is 3 with a rainbow color table.
cdo gmtcells temp > data.gmt makecpt -T213/318/3 -Crainbow > gmt.cpt psxy -K -JQ0/10i -Rd -L -Cgmt.cpt -m data.gmt > gmtplot.ps pscoast -O -J -R -Dc -W -B40g20 >> gmtplot.ps
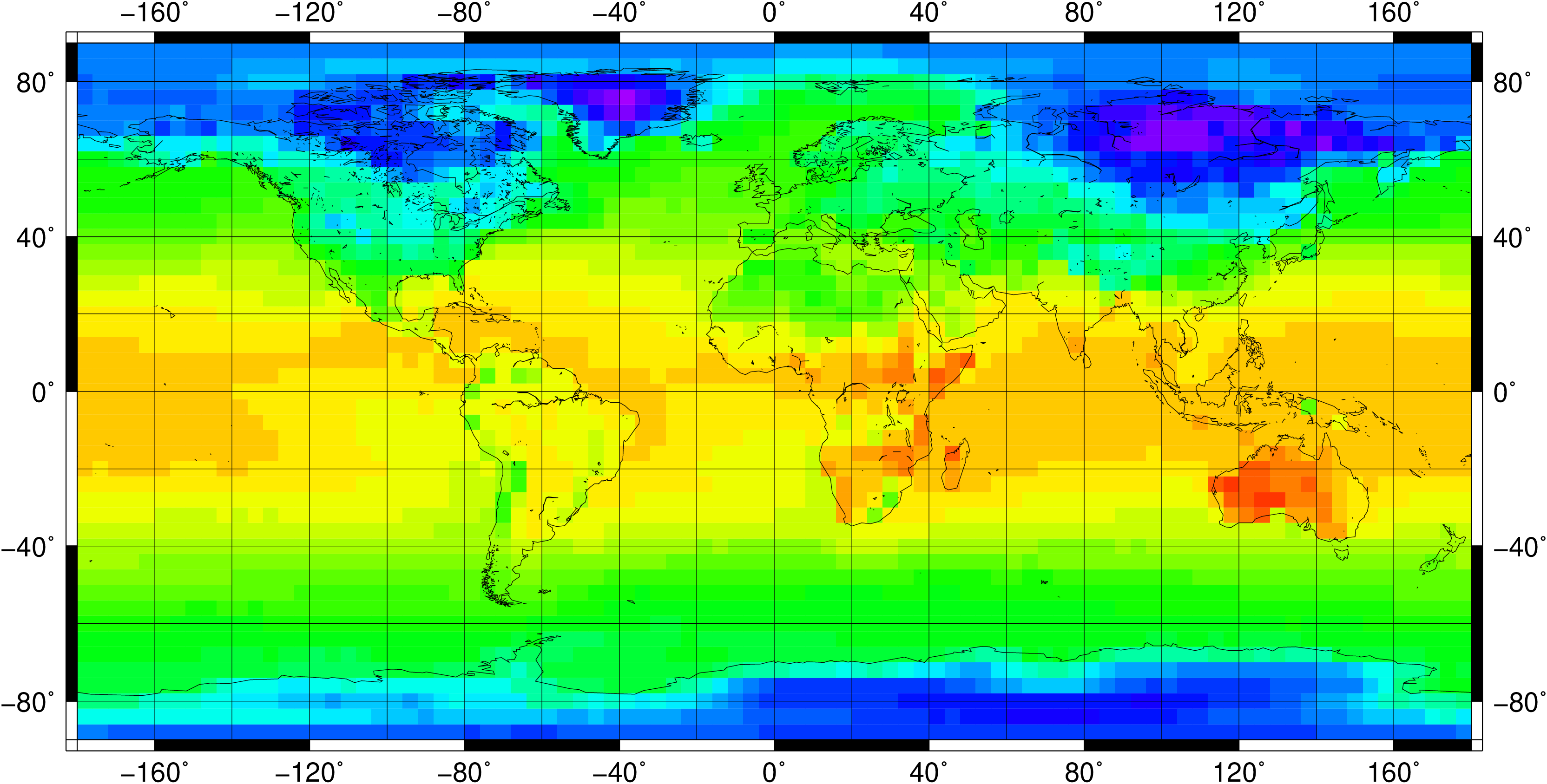
2.15 Miscellaneous
This section contains miscellaneous modules which do not fit to the other sections before.
Here is a short overview of all operators in this section:
| gradsdes | GrADS data descriptor file |
| after | ECHAM standard post processor |
| bandpass | Bandpass filtering |
| lowpass | Lowpass filtering |
| highpass | Highpass filtering |
| gridarea | Grid cell area |
| gridweights | Grid cell weights |
| smooth | Smooth grid points |
| smooth9 | 9 point smoothing |
| setvals | Set list of old values to new values |
| setrtoc | Set range to constant |
| setrtoc2 | Set range to constant others to constant2 |
| gridcellindex | Get grid cell index from lon/lat point |
| const | Create a constant field |
| random | Create a field with random numbers |
| topo | Create a field with topography |
| seq | Create a time series |
| stdatm | Create values for pressure and temperature for hydrostatic atmosphere |
| timsort | Sort over the time |
| uvDestag | Destaggering of u/v wind components |
| rotuvNorth | Rotate u/v wind to North pole. |
| projuvLatLon | Cylindrical Equidistant projection |
| rotuvb | Backward rotation |
| mrotuvb | Backward rotation of MPIOM data |
| mastrfu | Mass stream function |
| pressure_half | Pressure on half-levels |
| pressure | Pressure on full-levels |
| delta_pressure | Pressure difference of half-levels |
| sealevelpressure | Sea level pressure |
| gheight | Geopotential height on full-levels |
| gheight_half | Geopotential height on half-levels |
| air_density | Air density |
| adisit | Potential temperature to in-situ temperature |
| adipot | In-situ temperature to potential temperature |
| rhopot | Calculates potential density |
| histcount | Histogram count |
| histsum | Histogram sum |
| histmean | Histogram mean |
| histfreq | Histogram frequency |
| sethalo | Set the bounds of a field |
| wct | Windchill temperature |
| fdns | Frost days where no snow index per time period |
| strwin | Strong wind days index per time period |
| strbre | Strong breeze days index per time period |
| strgal | Strong gale days index per time period |
| hurr | Hurricane days index per time period |
| cmorlite | CMOR lite |
| verifygrid | Verify grid coordinates |
| hpdegrade | Degrade healpix |
| hpupgrade | Upgrade healpix |
2.15.1 GRADSDES - GrADS data descriptor file
Synopsis
gradsdes[,mapversion] infile
Description
Creates a [GrADS] data descriptor file. Supported file formats are GRIB1, NetCDF, SERVICE, EXTRA and IEG. For GRIB1 files the GrADS map file is also generated. For SERVICE and EXTRA files the grid have to be specified with the CDO option ’-g <grid>’. This module takes infile in order to create filenames for the descriptor (infile.ctl) and the map (infile.gmp) file.
Parameter
- mapversion
-
INTEGER Format version of the GrADS map file for GRIB1 datasets. Use 1 for a machine specific version 1 GrADS map file, 2 for a machine independent version 2 GrADS map file and 4 to support GRIB files >2GB. A version 2 map file can be used only with GrADS version 1.8 or newer. A version 4 map file can be used only with GrADS version 2.0 or newer. The default is 4 for files >2GB, otherwise 2.
Example
To create a GrADS data descriptor file from a GRIB1 dataset use:
cdo gradsdes infile.grb
This will create a descriptor file with the name infile.ctl and the map file infile.gmp.
Assumed the input GRIB1 dataset has 3 variables over 12 timesteps on a regular Gaussian F16 grid. The contents of the resulting GrADS data description file is approximately:
DSET ^infile.grb DTYPE GRIB INDEX ^infile.gmp XDEF 64 LINEAR 0.000000 5.625000 YDEF 32 LEVELS -85.761 -80.269 -74.745 -69.213 -63.679 -58.143 -52.607 -47.070 -41.532 -35.995 -30.458 -24.920 -19.382 -13.844 -8.307 -2.769 2.769 8.307 13.844 19.382 24.920 30.458 35.995 41.532 47.070 52.607 58.143 63.679 69.213 74.745 80.269 85.761 ZDEF 4 LEVELS 925 850 500 200 TDEF 12 LINEAR 12:00Z1jan1987 1mo TITLE infile.grb T21 grid OPTIONS yrev UNDEF -9e+33 VARS 3 geosp 0 129,1,0 surface geopotential (orography) [m^2/s^2] t 4 130,99,0 temperature [K] tslm1 0 139,1,0 surface temperature of land [K] ENDVARS
2.15.2 AFTERBURNER - ECHAM standard post processor
Synopsis
after[,vct] infiles outfile
Description
The "afterburner" is the standard post processor for [ECHAM] GRIB and NetCDF data which provides the following operations:
-
Extract specified variables and levels
-
Compute derived variables
-
Transform spectral data to Gaussian grid representation
-
Vertical interpolation to pressure levels
-
Compute temporal means
This operator reads selection parameters as namelist from stdin. Use the UNIX redirection "<namelistfile" to read the namelist from file.
The input files can’t be combined with other CDO operators because of an optimized reader for this operator.
Namelist
Namelist parameter and there defaults:
TYPE=0, CODE=-1, LEVEL=-1, INTERVAL=0, MEAN=0, EXTRAPOLATE=1
TYPE controls the transformation and vertical interpolation. Transforming spectral data to Gaussian grid representation and vertical interpolation to pressure levels are performed in a chain of steps. The TYPE parameter may be used to stop the chain at a certain step. Valid values are:
TYPE = 0 : Hybrid level spectral coefficients TYPE = 10 : Hybrid level fourier coefficients TYPE = 11 : Hybrid level zonal mean sections TYPE = 20 : Hybrid level gauss grids TYPE = 30 : Pressure level gauss grids TYPE = 40 : Pressure level fourier coefficients TYPE = 41 : Pressure level zonal mean sections TYPE = 50 : Pressure level spectral coefficients TYPE = 60 : Pressure level fourier coefficients TYPE = 61 : Pressure level zonal mean sections TYPE = 70 : Pressure level gauss grids
Vorticity, divergence, streamfunction and velocity potential need special treatment in the vertical transformation. They are not available as types 30, 40 and 41. If you select one of these combinations, type is automatically switched to the equivalent types 70, 60 and 61. The type of all other variables will be switched too, because the type is a global parameter.
CODE selects the variables by the ECHAM GRIB1 code number (1-255). The default value -1 processes all detected codes. Derived variables computed by the afterburner:
| Code | Name | Longname | Units | Level | Needed Codes |
| 34 | low_cld | low cloud | single | 223 on modellevel | |
| 35 | mid_cld | mid cloud | single | 223 on modellevel | |
| 36 | hih_cld | high cloud | single | 223 on modellevel | |
| 131 | u | u-velocity | m/s | atm (ml+pl) | 138, 155 |
| 132 | v | v-velocity | m/s | atm (ml+pl) | 138, 155 |
| 135 | omega | vertical velocity | Pa/s | atm (ml+pl) | 138, 152, 155 |
| 148 | stream | streamfunction | m 2/s 2/s | atm (ml+pl) | 131, 132 |
| 149 | velopot | velocity potential | m 2/s 2/s | atm (ml+pl) | 131, 132 |
| 151 | slp | mean sea level pressure | Pa | surface | 129, 130, 152 |
| 156 | geopoth | geopotential height | m | atm (ml+pl) | 129, 130, 133, 152 |
| 157 | rhumidity | relative humidity | atm (ml+pl) | 130, 133, 152 | |
| 189 | sclfs | surface solar cloud forcing | surface | 176-185 | |
| 190 | tclfs | surface thermal cloud forcing | surface | 177-186 | |
| 191 | sclf0 | top solar cloud forcing | surface | 178-187 | |
| 192 | tclf0 | top thermal cloud forcing | surface | 179-188 | |
| 259 | windspeed | windspeed | m/s | atm (ml+pl) | sqrt(u*u+v*v) |
| 260 | precip | total precipitation | surface | 142+143 | |
LEVEL selects the hybrid or pressure levels. The allowed values depends on the parameter TYPE. The default value -1 processes all detected levels.
INTERVAL selects the processing interval. The default value 0 process data on monthly intervals. INTERVAL=1 sets the interval to daily.
MEAN=1 compute and write monthly or daily mean fields. The default value 0 writes out all timesteps.
EXTRAPOLATE=0 switch of the extrapolation of missing values during the interpolation from model to pressure level (only available with MEAN=0 and TYPE=30). The default value 1 extrapolate missing values.
Possible combinations of TYPE, CODE and MEAN:
| TYPE | CODE | MEAN |
| 0/10/11 | 130 temperature | 0 |
| 0/10/11 | 131 u-velocity | 0 |
| 0/10/11 | 132 v-velocity | 0 |
| 0/10/11 | 133 specific humidity | 0 |
| 0/10/11 | 138 vorticity | 0 |
| 0/10/11 | 148 streamfunction | 0 |
| 0/10/11 | 149 velocity potential | 0 |
| 0/10/11 | 152 LnPs | 0 |
| 0/10/11 | 155 divergence | 0 |
| >11 | all codes | 0/1 |
Parameter
- vct
-
STRING File with VCT in ASCII format
Example
To interpolate ECHAM hybrid model level data to pressure levels of 925, 850, 500 and 200 hPa, use:
cdo after infile outfile << EON TYPE=30 LEVEL=92500,85000,50000,20000 EON
2.15.3 FILTER - Time series filtering
Synopsis
bandpass,fmin,fmax infile outfile
lowpass,fmax infile outfile
highpass,fmin infile outfile
Description
This module takes the time series for each gridpoint in infile and (fast fourier) transforms it into the frequency domain. According to the particular operator and its parameters certain frequencies are filtered (set to zero) in the frequency domain and the spectrum is (inverse fast fourier) transformed back into the time domain. To determine the frequency the time-axis of infile is used. (Data should have a constant time increment since this assumption applies for transformation. However, the time increment has to be different from zero.) All frequencies given as parameter are interpreted per year. This is done by the assumption of a 365-day calendar. Consequently if you want to perform multiyear-filtering accurately you have to delete the 29th of February. If your infile has a 360 year calendar the frequency parameters fmin respectively fmax should be multiplied with a factor of 360/365 in order to obtain accurate results. For the set up of a frequency filter the frequency parameters have to be adjusted to a frequency in the data. Here fmin is rounded down and fmax is always rounded up. Consequently it is possible to use bandpass with fmin=fmax without getting a zero-field for outfile. Hints for efficient usage:
-
to get reliable results the time-series has to be detrended (cdo detrend)
-
the lowest frequency greater zero that can be contained in infile is 1/(N*dT),
-
the greatest frequency is 1/(2dT) (Nyquist frequency),
with N the number of timesteps and dT the time increment of infile in years.
Missing value support for operators in this module is not implemented, yet!
Operators
- bandpass
-
Bandpass filtering
Bandpass filtering (pass for frequencies between fmin and fmax). Suppresses all variability outside the frequency range specified by [fmin,fmax]. - lowpass
-
Lowpass filtering
Lowpass filtering (pass for frequencies lower than fmax). Suppresses all variability with frequencies greater than fmax. - highpass
-
Highpass filtering
Highpass filtering (pass for frequencies greater than fmin). Suppresses all variabilty with frequencies lower than fmin.
Parameter
- fmin
-
FLOAT Minimum frequency per year that passes the filter.
- fmax
-
FLOAT Maximum frequency per year that passes the filter.
Note
For better performace of these operators use the CDO configure option --with-fftw3.
Example
Now assume your data are still hourly for a time period of 5 years but with a 365/366-day- calendar and you want to suppress the variability on timescales greater or equal to one year (we suggest here to use a number x bigger than one (e.g. x=1.5) since there will be dominant frequencies around the peak (if there is one) as well due to the issue that the time series is not of infinite length). Therefor you can use the following:
cdo highpass,x -del29feb infile outfile
Accordingly you might use the following to suppress variability on timescales shorter than one year:
cdo lowpass,1 -del29feb infile outfile
Finally you might be interested in 2-year variability. If you want to suppress the seasonal cycle as well as say the longer cycles in climate system you might use
cdo bandpass,x,y -del29feb infile outfile
with x<=0.5 and y >=0.5.
2.15.4 GRIDCELL - Grid cell quantities
Synopsis
gridarea[,radius] infile outfile
gridweights infile outfile
Description
This module reads the grid cell area of the first grid from the input stream. If the grid cell area is missing it will be computed from the grid coordinates. The area of a grid cell is calculated using spherical triangles from the coordinates of the center and the vertices. The base is a unit sphere which is scaled with the radius of the planet. The default planet radius is 6371000 meter. The parameter radius or the environment variable PLANET_RADIUS can be used to change the default. Depending on the chosen operator the grid cell area or weights are written to the output stream.
Operators
- gridarea
-
Grid cell area
Writes the grid cell area to the output stream. If the grid cell area have to be computed it is scaled with the planet radius to square meters. - gridweights
-
Grid cell weights
Writes the grid cell area weights to the output stream.
Parameter
- radius
-
FLOAT Planet radius in meter
Environment
- PLANET_RADIUS
-
This variable is used to scale the computed grid cell areas to square meters. By default PLANET_RADIUS is set to an earth radius of 6371000 meter.
2.15.5 SMOOTH - Smooth grid points
Synopsis
smooth[,options] infile outfile
smooth9 infile outfile
Description
Smooth all grid points of a horizontal grid. Options is a comma-separated list of "key=value" pairs with optional parameters.
Operators
- smooth
-
Smooth grid points
Performs a N point smoothing on all input fields. The number of points used depend on the search radius (radius) and the maximum number of points (maxpoints). Per default all points within the search radius of 1degree are used. The weights for the points depend on the weighting method and the distance. The implemented weighting method is linear with constant default weights of 0.25 at distance 0 (weight0) and at the search radius (weightR). - smooth9
-
9 point smoothing
Performs a 9 point smoothing on all fields with a quadrilateral curvilinear grid. The result at each grid point is a weighted average of the grid point plus the 8 surrounding points. The center point receives a weight of 1.0, the points at each side and above and below receive a weight of 0.5, and corner points receive a weight of 0.3. All 9 points are multiplied by their weights and summed, then divided by the total weight to obtain the smoothed value. Any missing data points are not included in the sum; points beyond the grid boundary are considered to be missing. Thus the final result may be the result of an averaging with less than 9 points.
Parameter
- nsmooth
-
INTEGER Number of times to smooth, default nsmooth=1
- radius
-
STRING Search radius, default radius=1deg (units: deg, rad, km, m)
- maxpoints
-
INTEGER Maximum number of points, default maxpoints=<gridsize>
- weighted
-
STRING Weighting method, default weighted=linear
- weight0
-
FLOAT Weight at distance 0, default weight0=0.25
- weightR
-
FLOAT Weight at the search radius, default weightR=0.25
2.15.6 DELTAT - Difference between timesteps
Synopsis
deltat infile outfile
Description
This operator computes the difference between each timestep.
2.15.7 REPLACEVALUES - Replace variable values
Synopsis
setvals,oldval,newval[,...] infile outfile
setrtoc,rmin,rmax,c infile outfile
setrtoc2,rmin,rmax,c,c2 infile outfile
Description
This module replaces old variable values with new values, depending on the operator.
Operators
- setvals
-
Set list of old values to new values
Supply a list of n pairs of old and new values. - setrtoc
-
Set range to constant
o(t,x) =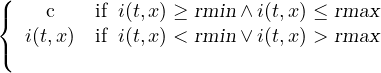
- setrtoc2
-
Set range to constant others to constant2
o(t,x) =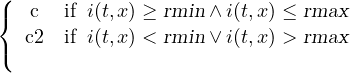
Parameter
- oldval,newval,...
-
FLOAT Pairs of old and new values
- rmin
-
FLOAT Lower bound
- rmax
-
FLOAT Upper bound
- c
-
FLOAT New value - inside range
- c2
-
FLOAT New value - outside range
2.15.8 GETGRIDCELL - Get grid cell index
Synopsis
gridcellindex[,parameter] infile
Description
Get the grid cell index of one grid point selected by the parameter lon and lat.
Parameter
- lon
-
INTEGER Longitude of the grid cell in degree
- lat
-
INTEGER Latitude of the grid cell in degree
2.15.9 VARGEN - Generate a field
Synopsis
const,const,grid outfile
random,grid[,seed] outfile
topo[,grid] outfile
seq,start,end[,inc] outfile
stdatm,levels outfile
Description
Generates a dataset with one or more fields
Operators
- const
-
Create a constant field
Creates a constant field. All field elements of the grid have the same value. - random
-
Create a field with random numbers
Creates a field with rectangularly distrubuted random numbers in the interval [0,1]. - topo
-
Create a field with topography
Creates a field with topography data, per default on a global half degree grid. - seq
-
Create a time series
Creates a time series with field size 1 and field elements beginning with a start value in time step 1 which is increased from one time step to the next. - stdatm
-
Create values for pressure and temperature for hydrostatic atmosphere
Creates pressure and temperature values for the given list of vertical levels. The formulas are:P(z) = P0 exp
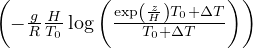
T(z) = T0 + ΔT exp
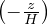
with the following constants

This is the solution for the hydrostatic equations and is only valid for the troposphere (constant positive lapse rate). The temperature increase in the stratosphere and other effects of the upper atmosphere are not taken into account.
Parameter
- const
-
FLOAT Constant
- seed
-
INTEGER The seed for a new sequence of pseudo-random numbers [default: 1]
- grid
-
STRING Target grid description file or name
- start
-
FLOAT Start value of the loop
- end
-
FLOAT End value of the loop
- inc
-
FLOAT Increment of the loop [default: 1]
- levels
-
FLOAT Target levels in metre above surface
Example
To create a standard atmosphere dataset on a given horizontal grid:
cdo enlarge,gridfile -stdatm,10000,8000,5000,3000,2000,1000,500,200,0 outfile
2.15.10 TIMSORT - Timsort
Synopsis
timsort infile outfile
Description
Sorts the elements in ascending order over all timesteps for every field position. After sorting it is:
o(t1,x) <= o(t2,x) ∀(t1 < t2),x
Example
To sort all field elements of a dataset over all timesteps use:
cdo timsort infile outfile
2.15.11 WINDTRANS - Wind Transformation
Synopsis
uvDestag,u,v[,-/+0.5[,-/+0.5]] infile outfile
rotuvNorth,u,v infile outfile
projuvLatLon,u,v infile outfile
Description
This module contains special operators for datsets with wind components on a rotated lon/lat grid, e.g. data from the regional model HIRLAM or REMO.
Operators
- uvDestag
-
Destaggering of u/v wind components
This is a special operator for destaggering of wind components. If the file contains a grid with temperature (name=’t’ or code=11) then grid_temp will be used for destaggered wind. - rotuvNorth
-
Rotate u/v wind to North pole.
This is an operator for transformation of wind-vectors from grid-relative to north-pole relative for the whole file. (FAST implementation with JACOBIANS) - projuvLatLon
-
Cylindrical Equidistant projection
Thus is an operator for transformation of wind-vectors from the globe-spherical coordinate system into a flat Cylindrical Equidistant (lat-lon) projection. (FAST JACOBIAN implementation)
Parameter
- u,v
-
STRING Pair of u,v wind components (use variable names or code numbers)
- -/+0.5,-/+0.5
-
STRING Destaggered grid offsets are optional (default -0.5,-0.5)
Example
Typical operator sequence on HIRLAM NWP model output (LAMH_D11 files):
cdo uvDestag,33,34 inputfile inputfile_destag cdo rotuvNorth,33,34 inputfile_destag inputfile_rotuvN
2.15.12 ROTUV - Rotation
Synopsis
rotuvb,u,v,... infile outfile
Description
This is a special operator for datsets with wind components on a rotated grid, e.g. data from the regional model REMO. It performs a backward transformation of velocity components U and V from a rotated spherical system to a geographical system.
Parameter
- u,v,...
-
STRING Pairs of zonal and meridional velocity components (use variable names or code numbers)
Note
This is a specific implementation for data from the REMO model, it may not work with data from other sources.
Example
To transform the u and v velocity of a dataset from a rotated spherical system to a geographical system use:
cdo rotuvb,u,v infile outfile
2.15.13 MROTUVB - Backward rotation of MPIOM data
Synopsis
mrotuvb infile1 infile2 outfile
Description
MPIOM data are on a rotated Arakawa C grid. The velocity components U and V are located on the edges of the cells and point in the direction of the grid lines and rows. With mrotuvb the velocity vector is rotated in latitudinal and longitudinal direction. Before the rotation, U and V are interpolated to the scalar points (cell center). U is located with the coordinates for U in infile1 and V in infile2. mrotuvb assumes a positive meridional flow for a flow from grid point(i,j) to grid point(i,j+1) and positive zonal flow for a flow from grid point(i+1,j) to point(i,j).
Note
This is a specific implementation for data from the MPIOM model, it may not work with data from other sources.
2.15.14 MASTRFU - Mass stream function
Synopsis
mastrfu infile outfile
Description
This is a special operator for the post processing of the atmospheric general circulation model [ECHAM]. It computes the mass stream function (code=272). The input dataset have to be a zonal mean of v-velocity [m/s] (code=132) on pressure levels.
Example
To compute the mass stream function from a zonal mean v-velocity dataset use:
cdo mastrfu infile outfile
2.15.15 PRESSURE - Pressure on model levels
Synopsis
<operator> infile outfile
Description
This module contains operators to calculate the pressure on model levels. To calculate the pressure on model levels, the a and b coefficients defining the model levels and the surface pressure are required. The a and b coefficients are normally part of the model level data. If not available, the surface pressure can be derived from the logarithm of the surface pressure. The surface pressure is identified by the GRIB1 code number or NetCDF CF standard name.
| Name | Units | GRIB1 code | CF standard name |
| log surface pressure | Pa | 152 | |
| surface pressure | Pa | 134 | surface_air_pressure |
Operators
- pressure_half
-
Pressure on half-levels
This operator computes the pressure on model half-levels in pascal. The model half-level pressure (p_half) is given by: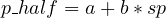
with
a,b: coefficients defining the model levels
sp: surface pressure - pressure
-
Pressure on full-levels
This operator computes the pressure on model full-levels in pascal. The pressure on model full-levels (p_full) is in the middle of the layers defined by the model half-levels: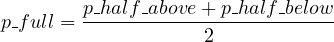
- delta_pressure
-
Pressure difference of half-levels
This operator computes the pressure difference between to model half-levels.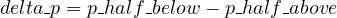
2.15.16 DERIVEPAR - Derived model parameters
Synopsis
<operator> infile outfile
Description
This module contains operators that calculate derived model parameters. All necessary input variables are identified by their GRIB1 code number or the NetCDF CF standard name. Supported GRIB1 parameter tables are: WMO standard table number 2 and ECMWF local table number 128.
| CF standard name | Units | GRIB 1 code |
| surface_air_pressure | Pa | 134 |
| air_temperature | K | 130 |
| specific_humidity | kg/kg | 133 |
| surface_geopotential | m2 s-2 | 129 |
| geopotential_height | m | 156 |
Operators
- sealevelpressure
-
Sea level pressure
This operator computes the sea level pressure (air_pressure_at_sea_level). Required input fields are surface_air_pressure, surface_geopotential and air_temperature on full hybrid sigma pressure levels. - gheight
-
Geopotential height on full-levels
This operator computes the geopotential height (geopotential_height) on model full-levels in metres. Required input fields are surface_air_pressure, surface_geopotential, specific_humidity and air_temperature on full hybrid sigma pressure levels. Note, this procedure is an approximation, which doesn’t take into account the effects of e.g. cloud ice and water, rain and snow. - gheight_half
-
Geopotential height on half-levels
This operator computes the geopotential height (geopotential_height) on model half-levels in metres. Required input fields are surface_air_pressure, surface_geopotential, specific_humidity and air_temperature on full hybrid sigma pressure levels. Note, this procedure is an approximation, which doesn’t take into account the effects of e.g. cloud ice and water, rain and snow. - air_density
-
Air density
This operator computes the air density, it depends on pressure, humidity and temperature. Required input fields are surface_air_pressure, specific_humidity and air_temperature on full hybrid sigma pressure levels. The air density (rho) is calculated with the following formula: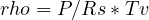
P: air pressure in Pascal
Tv: virtual temperature in Kelvin
Rs: specific gas constant for try air; 287.085 J/(kg*K)![Tv = T * [1 + a*q]](cdo186x.png)
T: air temperature in Kelvin
q: specific humidity
a: gas constants of air and water vapor; 0.6078
2.15.17 ADISIT - Potential temperature to in-situ temperature and vice versa
Synopsis
<operator>[,pressure] infile outfile
Description
Operators
- adisit
-
Potential temperature to in-situ temperature
This is a special operator for the post processing of the ocean and sea ice model [MPIOM]. It converts potential temperature adiabatically to in-situ temperature to(t, s, p). Required input fields are sea water potential temperature (name=tho; code=2) and sea water salinity (name=sao; code=5). Pressure is calculated from the level information or can be specified by the optional parameter. Output fields are sea water temperature (name=to; code=20) and sea water salinity (name=s; code=5). - adipot
-
In-situ temperature to potential temperature
This is a special operator for the post processing of the ocean and sea ice model [MPIOM]. It converts in-situ temperature to potential temperature tho(to, s, p). Required input fields are sea water in-situ temperature (name=t; code=2) and sea water salinity (name=sao,s; code=5). Pressure is calculated from the level information or can be specified by the optional parameter. Output fields are sea water temperature (name=tho; code=2) and sea water salinity (name=s; code=5).
Parameter
- pressure
-
FLOAT Pressure in bar (constant value assigned to all levels)
2.15.18 RHOPOT - Calculates potential density
Synopsis
rhopot[,pressure] infile outfile
Description
This is a special operator for the post processing of the ocean and sea ice model [MPIOM]. It calculates the sea water potential density (name=rhopoto; code=18). Required input fields are sea water in-situ temperature (name=to; code=20) and sea water salinity (name=sao; code=5). Pressure is calculated from the level information or can be specified by the optional parameter.
Parameter
- pressure
-
FLOAT Pressure in bar (constant value assigned to all levels)
Example
To compute the sea water potential density from the potential temperature use this operator in combination with adisit:
cdo rhopot -adisit infile outfile
2.15.19 HISTOGRAM - Histogram
Synopsis
<operator>,bounds infile outfile
Description
This module creates bins for a histogram of the input data. The bins have to be adjacent and have non-overlapping intervals. The user has to define the bounds of the bins. The first value is the lower bound and the second value the upper bound of the first bin. The bounds of the second bin are defined by the second and third value, aso. Only 2-dimensional input fields are allowed. The output file contains one vertical level for each of the bins requested.
Operators
- histcount
-
Histogram count
Number of elements in the bin range. - histsum
-
Histogram sum
Sum of elements in the bin range. - histmean
-
Histogram mean
Mean of elements in the bin range. - histfreq
-
Histogram frequency
Relative frequency of elements in the bin range.
Parameter
- bounds
-
FLOAT Comma-separated list of the bin bounds (-inf and inf valid)
2.15.20 SETHALO - Set the bounds of a field
Synopsis
sethalo[,parameter] infile outfile
Description
This operator sets the boundary in the east, west, south and north of the rectangular understood fields. Positive values of the parameters increase the boundary in the selected direction. Negative values decrease the field at the selected boundary. The new rows and columns are filled with the missing value. With the optional parameter value a different fill value can be used. Global cyclic fields are filled cyclically at the east and west borders, if the fill value is not set by the user. All input fields need to have the same horizontal grid.
Parameter
- east
-
INTEGER East halo
- west
-
INTEGER West halo
- south
-
INTEGER South halo
- north
-
INTEGER North halo
- value
-
FLOAT Fill value (default is the missing value)
2.15.21 WCT - Windchill temperature
Synopsis
wct infile1 infile2 outfile
Description
Let infile1 and infile2 be time series of temperature and wind speed fields, then a corresponding time series of resulting windchill temperatures is written to outfile. The wind chill temperature calculation is only valid for a temperature of T <= 33 ℃ and a wind speed of v >= 1.39 m/s. Whenever these conditions are not satisfied, a missing value is written to outfile. Note that temperature and wind speed fields have to be given in units of ℃ and m/s, respectively.
2.15.22 FDNS - Frost days where no snow index per time period
Synopsis
fdns infile1 infile2 outfile
Description
Let infile1 be a time series of the daily minimum temperature TN and infile2 be a corresponding series of daily surface snow amounts. Then the number of days where TN < 0 ℃ and the surface snow amount is less than 1 cm is counted. The temperature TN have to be given in units of Kelvin. The date information of a timestep in outfile is the date of the last contributing timestep in infile.
2.15.23 STRWIN - Strong wind days index per time period
Synopsis
strwin[,v] infile outfile
Description
Let infile be a time series of the daily maximum horizontal wind speed VX, then the number of days where VX > v is counted. The horizontal wind speed v is an optional parameter with default v = 10.5 m/s. A further output variable is the maximum number of consecutive days with maximum wind speed greater than or equal to v. Note that both VX and v have to be given in units of m/s. Also note that the horizontal wind speed is defined as the square root of the sum of squares of the zonal and meridional wind speeds. The date information of a timestep in outfile is the date of the last contributing timestep in infile.
Parameter
- v
-
FLOAT Horizontal wind speed threshold (m/s, default v = 10.5 m/s)
2.15.24 STRBRE - Strong breeze days index per time period
Synopsis
strbre infile outfile
Description
Let infile be a time series of the daily maximum horizontal wind speed VX, then the number of days where VX is greater than or equal to 10.5 m/s is counted. A further output variable is the maximum number of consecutive days with maximum wind speed greater than or equal to 10.5 m/s. Note that VX is defined as the square root of the sum of squares of the zonal and meridional wind speeds and have to be given in units of m/s. The date information of a timestep in outfile is the date of the last contributing timestep in infile.
2.15.25 STRGAL - Strong gale days index per time period
Synopsis
strgal infile outfile
Description
Let infile be a time series of the daily maximum horizontal wind speed VX, then the number of days where VX is greater than or equal to 20.5 m/s is counted. A further output variable is the maximum number of consecutive days with maximum wind speed greater than or equal to 20.5 m/s. Note that VX is defined as the square root of the sum of square of the zonal and meridional wind speeds and have to be given in units of m/s. The date information of a timestep in outfile is the date of the last contributing timestep in infile.
2.15.26 HURR - Hurricane days index per time period
Synopsis
hurr infile outfile
Description
Let infile be a time series of the daily maximum horizontal wind speed VX, then the number of days where VX is greater than or equal to 32.5 m/s is counted. A further output variable is the maximum number of consecutive days with maximum wind speed greater than or equal to 32.5 m/s. Note that VX is defined as the square root of the sum of squares of the zonal and meridional wind speeds and have to be given in units of m/s. The date information of a timestep in outfile is the date of the last contributing timestep in infile.
2.15.27 CMORLITE - CMOR lite
Synopsis
cmorlite,table[,convert] infile outfile
Description
The [CMOR] (Climate Model Output Rewriter) library comprises a set of functions, that can be used to produce CF-compliant NetCDF files that fulfill the requirements of many of the climate community’s standard model experiments. These experiments are collectively referred to as MIP’s. Much of the metadata written to the output files is defined in MIP-specific tables, typically made available from each MIP’s web site.
The CDO operator cmorlite process the header and variable section of such MIP tables and writes the result with the internal IO library [CDI]. In addition to the CMOR 2 and 3 table format, the CDO parameter table format is also supported. The following parameter table entries are available:
| Entry | Type | Description |
| name | WORD | Name of the variable |
| out_name | WORD | New name of the variable |
| type | WORD | Data type (real or double) |
| standard_name | WORD | As defined in the CF standard name table |
| long_name | STRING | Describing the variable |
| units | STRING | Specifying the units for the variable |
| comment | STRING | Information concerning the variable |
| cell_methods | STRING | Information concerning calculation of means or climatologies |
| cell_measures | STRING | Indicates the names of the variables containing cell areas and volumes |
| missing_value | FLOAT | Specifying how missing data will be identified |
| valid_min | FLOAT | Minimum valid value |
| valid_max | FLOAT | Maximum valid value |
| ok_min_mean_abs | FLOAT | Minimum absolute mean |
| ok_max_mean_abs | FLOAT | Maximum absolute mean |
| factor | FLOAT | Scale factor |
| delete | INTEGER | Set to 1 to delete variable |
| convert | INTEGER | Set to 1 to convert the unit if necessary |
Most of the above entries are stored as variables attributes, some of them are handled differently. The variable name is used as a search key for the parameter table. valid_min, valid_max, ok_min_mean_abs and ok_max_mean_abs are used to check the range of the data.
Parameter
- table
-
STRING Name of the CMOR table as specified from PCMDI
- convert
-
STRING Converts the units if necessary
Example
Here is an example of a parameter table for one variable:
prompt> cat mypartab ¶meter name = t out_name = ta standard_name = air_temperature units = "K" missing_value = 1.0e+20 valid_min = 157.1 valid_max = 336.3 /
To apply this parameter table to a dataset use:
cdo -f nc cmorlite,mypartab,convert infile outfile
This command renames the variable t to ta. The standard name of this variable is set to air_temperature and the unit is set to [K] (converts the unit if necessary). The missing value will be set to 1.0e+20. In addition it will be checked whether the values of the variable are in the range of 157.1 to 336.3. The result will be stored in NetCDF.
2.15.28 VERIFYGRID - Verify grid coordinates
Synopsis
verifygrid infile
Description
This operator verifies the coordinates of all horizontal grids found in infile. Among other things, it searches for duplicate cells, non-convex cells, and whether the center is located outside the cell bounds. Use the CDO option -v to output the position of these cells. This information can be useful to avoid problems when interpolating the data.
2.15.29 HEALPIX - Change healpix resolution
Synopsis
<operator>,parameter infile outfile
Description
Degrade or upgrade the resolution of a healpix grid.
Operators
- hpdegrade
-
Degrade healpix
Degrade the resolution of a healpix grid. The value of the target pixel is the mean of the source pixels. - hpupgrade
-
Upgrade healpix
Upgrade the resolution of a healpix grid. The values of the target pixels are the value of the source pixel.
Parameter
- nside
-
INTEGER The nside of the target healpix, must be a power of two [default: same as input].
- order
-
STRING Pixel ordering of the target healpix (’nested’ or ’ring’).
- power
-
FLOAT If non-zero, divide the result by (nside[in]/nside[out])**power. power=-2 keeps the sum of the map invariant.
3 Contributors
3.1 History
CDO was originally developed by Uwe Schulzweida at the Max Planck Institute for Meteorology (MPI-M). The first public release is available since 2003. The MPI-M, together with the DKRZ, has a long history in the development of tools for processing climate data. CDO was inspired by some of these tools, such as the PINGO package and the GRIB-Modules.
PINGO1 was developed by Juergen Waszkewitz, Peter Lenzen, and Nathan Gillet in 1995 at the DKRZ, Hamburg (Germany). CDO has a similar user interface and uses some of the PINGO routines.
The GRIB-Modules was developed by Heiko Borgert and Wolfgang Welke in 1991 at the MPI-M. CDO is using a similar module structure and also some of the routines.
3.2 External sources
CDO has incorporated code from several sources:
- afterburner
-
is a postprocessing application for ECHAM data and ECMWF analysis data, originally developed by Edilbert Kirk, Michael Ponater and Arno Hellbach. The afterburner code was modified for the CDO operators after, ml2pl, ml2hl, sp2gp, gp2sp.
- SCRIP
-
is a software package used to generate interpolation weights for remapping fields from one grid to another in spherical geometry [SCRIP]. It was developed at the Los Alamos National Laboratory by Philip W. Jones. The SCRIP library was converted from Fortran to ANSI C and is used as the base for the remapping operators in CDO.
- YAC
-
(Yet Another Coupler) was jointly developed by DKRZ and MPI-M by Moritz Hanke and Rene Redler [YAC]. CDO is using the clipping and cell search routines for the conservative remapping with remapcon.
- libkdtree
-
a C99 implementation of the kd-tree algorithm developed by Juerg Dietrich.
CDO uses tools from the GNU project, including automake, and libtool.
3.3 Contributors
The primary contributors to the CDO development have been:
- Uwe Schulweida
-
: Concept, design and implementation of CDO, project coordination, and releases.
- Luis Kornblueh
-
: He supports CDO from the beginning. His main contributions are GRIB performance and compression, GME and unstructured grid support. Luis also helps with design and planning.
- Ralf Mueller
-
: He is working on CDO since 2009. His main contributions are the implementation of the User Portal, the ruby and python interface for all CDO operators, the building process and the Windows support. The CDO User Portal was funded by the European Commission infracstructure project IS-ENES. Ralf also helps a lot with the user support. Implemented operators: intlevel3d, consecsum, consects, ngrids, ngridpoints, reducegrid
- Oliver Heidmann
-
: He is working on CDO since 2015. His main contributions are refactoring to C++ and the new command line parser.
- Karin Meier-Fleischer
-
: She is working in the CDO user support since 2017.
- Fabian Wachsmann
-
: He is working on CDO for the CMIP6 project since 2016. His main task is the implementation and support of the cmor operator. He has also implemented the ETCCDI Indices of Daily Temperature and Precipitation Extremes.
- Cedrick Ansorge
-
: He worked on the software package CDO as a student assistant at MPI-M from 2007-2011. Implemented operators: eof, eof3d, enscrps, ensbrs, maskregion, bandpass, lowpass, highpass, smooth9
- Ralf Quast
-
: He worked on CDO on behalf of the Service Gruppe Anpassung (SGA), DKRZ in 2006. He implemented all ECA Indices of Daily Temperature and Precipitation Extremes, all percentile operators, module YDRUNSTAT and wct.
- Kameswarrao Modali
-
: He worked on CDO from 2012-2013.
Implemented operators: contour, shaded, grfill, vector, graph. - Michal Koutek
-
: Implemented operators: selmulti delmulti, changemulti, samplegrid, uvDestag, rotuvNorth, projuvLatLon.
- Etienne Tourigny
-
: Implemented operators: setclonlatbox, setcindexbox, setvals, splitsel, histfreq, setrtoc, setrtoc2.
- Karl-Hermann Wieners
-
: Implemented operators: aexpr, aexprf, selzaxisname.
- Asela Rajapakse
-
: He worked on CDO from 2016-2017 as part of the EUDAT project.
Implemented operator: verifygrid - Estanislao Gavilan
-
: Improved the CDO documentation for the installation section.
Many users have contributed to CDO by sending bug reports, patches and suggestions over time. Very helpful is also the active participation in the user forum of some users. Here is an incomplete list:
Jaison-Thomas Ambadan, Harald Anlauf, Andy Aschwanden, Stefan Bauer, Simon Blessing, Renate Brokopf, Michael Boettinger, Tim Bruecher, Reinhard Budich, Martin Claus, Traute Crueger, Brendan de Tracey, Irene Fischer-Bruns, Chris Fletscher, Helmut Frank, Kristina Froehlich, Oliver Fuhrer, Monika Esch, Pier Giuseppe Fogli, Beate Gayer, Veronika Gayler, Marco Giorgetta, David Gobbett, Holger Goettel, Helmut Haak, Stefan Hagemann, Angelika Heil, Barbara Hennemuth, Daniel Hernandez, Nathanael Huebbe, Thomas Jahns, Frank Kaspar, Daniel Klocke, Edi Kirk, Yvonne Kuestermann, Stefanie Legutke, Leonidas Linardakis, Stephan Lorenz, Frank Lunkeit, Uwe Mikolajewicz, Laura Niederdrenk, Dirk Notz, Hans-Juergen Panitz, Ronny Petrik, Swantje Preuschmann, Florian Prill, Asela Rajapakse, Daniel Reinert, Hannes Reuter, Mathis Rosenhauer, Reiner Schnur, Martin Schultz, Dennis Shea, Kevin Sieck, Martin Stendel, Bjorn Stevens, Martina Stockhaus, Claas Teichmann, Adrian Tompkins, Joerg Trentmann, Alvaro M. Valdebenito, Geert Jan van Oldenborgh, Jin-Song von Storch, David Wang, Joerg Wegner, Heiner Widmann, Claudia Wunram, Klaus Wyser
Please let me know if your name was omitted!
Bibliography
[BitInformation.jl]
M Kloewer, M Razinger, JJ Dominguez, PD Dueben and TN Palmer, 2021. Compressing
atmospheric data into its real information content. Nature Computational Science 1, 713-724.
10.1038/s43588-021-00156-2
[CDI]
Climate Data Interface, from the Max Planck Institute for Meteorologie
[CM-SAF]
Satellite Application Facility on Climate Monitoring, from the German Weather Service (Deutscher
Wetterdienst, DWD)
[CMOR]
Climate Model Output Rewriter, from the Program For Climate Model Diagnosis and
Intercomparison (PCMDI)
[ecCodes]
API for GRIB decoding/encoding, from the European Centre for Medium-Range Weather Forecasts
( ECMWF)
[ECHAM]
The atmospheric general circulation model ECHAM5, from the Max Planck Institute for Meteorologie
[GMT]
The Generic Mapping Tool, from the School of Ocean and Earth Science and Technology ( SOEST)
[GrADS]
Grid Analysis and Display System, from the Center for Ocean-Land-Atmosphere Studies ( COLA)
[GRIB]
GRIB version 1, from the World Meteorological Organisation ( WMO)
[HDF5]
HDF version 5, from the HDF Group
[INTERA]
INTERA Software Package, from the Max Planck Institute for Meteorologie
[Magics]
Magics Software Package, from the European Centre for Medium-Range Weather Forecasts
(ECMWF)
[MPIOM]
Ocean and sea ice model, from the Max Planck Institute for Meteorologie
[NetCDF]
NetCDF Software Package, from the UNIDATA Program Center of the University Corporation for
Atmospheric Research
[PINGO]
The PINGO package, from the Model & Data group at the Max Planck Institute for Meteorologie
[REMO]
Regional Model, from the Max Planck Institute for Meteorologie
[Preisendorfer]
Rudolph W. Preisendorfer: Principal Component Analysis in Meteorology and Oceanography, Elsevier
(1988)
[PROJ]
Cartographic Projections Library, originally written by Gerald Evenden then of the USGS.
[SCRIP]
SCRIP Software Package, from the Los Alamos National Laboratory
[szip]
Szip compression software, developed at University of New Mexico.
[vonStorch]
Hans von Storch, Walter Zwiers: Statistical Analysis in Climate Research, Cambridge University Press
(1999)
[YAC]
YAC - Yet Another Coupler Software Package, from DKRZ and MPI for Meteorologie
A. Environment Variables
The following table describes the environment variables that affect CDO.
| Variable name | Default | Description |
| CDO_DOWNLOAD_PATH | None | Path where CDO can store downloads. |
| CDO_FILE_SUFFIX | None | Default filename suffix. This suffix will be added to the output file |
| name instead of the filename extension derived from the file | ||
| format. NULL will disable the adding of a file suffix. | ||
| CDO_GRIDSEARCH_RADIUS | 180 | Grid search radius in degree. Used by the operators |
| setmisstonn, remapdis and remapnn. | ||
| CDO_HISTORY_INFO | true | ’false’ don’t write information to the global history attribute. |
| CDO_ICON_GRIDS | None | Root directory of the installed ICON grids (e.g. /pool/data/ICON). |
| CDO_PCTL_NBINS | 101 | Number of histogram bins. |
| CDO_RESET_HISTORY | false | ’true’ resets the global history attribute. |
| CDO_REMAP_NORM | fracarea | Choose the normalization for the conservative interpolation |
| CDO_TIMESTAT_DATE | None | Set target timestamp of a temporal statistic operator to the "first", |
| "middle", "midhigh" or "last" contributing source timestep. | ||
| CDO_USE_FFTW | 1 | Set to 0 to switch off usage of FFTW. Used in the Filter module. |
| CDO_VERSION_INFO | true | ’false’ disables the global NetCDF attribute CDO. |
B. Parallelized operators
Some of the CDO operators are parallelized with OpenMP. To use CDO with multiple OpenMP threads, you have to set the number of threads with the option ’-P’. Here is an example to distribute the bilinear interpolation on 8 OpenMP threads:
cdo -P 8 remapbil,targetgrid infile outfile
The following CDO operators are parallelized with OpenMP:
| Module | Operator | Description |
| Afterburner | after | ECHAM standard post processor |
| Detrend | detrend | Detrend |
| EcaEtccdi | etccdi_tx90p | % of days when daily max temperature is > the 90th percentile |
| EcaEtccdi | etccdi_tx10p | % of days when daily max temperature is < the 10th percentile |
| EcaEtccdi | etccdi_tn90p | % of days when daily min temperature is > the 90th percentile |
| EcaEtccdi | etccdi_tn10p | % of days when daily min temperature is < the 10th percentile |
| EcaEtccdi | etccdi_r95p | Annual tot precip when daily precip exceeds the 95th percentile of ... |
| EcaEtccdi | etccdi_r99p | Annual tot precip when daily precip exceeds the 99th percentile of ... |
| Ensstat | ens<STAT> | Statistical values over an ensemble |
| EOF | eof | Empirical Orthogonal Functions |
| Fillmiss | setmisstonn | Set missing value to nearest neighbor |
| Fillmiss | setmisstodis | Set missing value to distance-weighted average |
| Filter | bandpass | Bandpass filtering |
| Filter | lowpass | Lowpass filtering |
| Filter | highpass | Highpass filtering |
| Fourier | fourier | Fourier transformation |
| Genweights | genbil | Generate bilinear interpolation weights |
| Genweights | genbic | Generate bicubic interpolation weights |
| Genweights | gendis | Generate distance-weighted average remap weights |
| Genweights | gennn | Generate nearest neighbor remap weights |
| Genweights | gencon | Generate 1st order conservative remap weights |
| Genweights | gencon2 | Generate 2nd order conservative remap weights |
| Genweights | genlaf | Generate largest area fraction remap weights |
| Gridboxstat | gridbox<STAT> | Statistical values over grid boxes |
| Intlevel | intlevel | Linear level interpolation |
| Intlevel3d | intlevel3d | Linear level interpolation from/to 3D vertical coordinates |
| Remapeta | remapeta | Remap vertical hybrid level |
| Remap | remapbil | Bilinear interpolation |
| Remap | remapbic | Bicubic interpolation |
| Remap | remapdis | Distance-weighted average remapping |
| Remap | remapnn | Nearest neighbor remapping |
| Remap | remapcon | First order conservative remapping |
| Remap | remaplaf | Largest area fraction remapping |
| Smooth | smooth | Smooth grid points |
| Spectral | sp2gp, gp2sp | Spectral transformation |
| Module | Operator | Description |
| Vertintap | ap2pl, ap2hl | Vertical interpolation on hybrid sigma height coordinates |
| Vertintgh | gh2hl | Vertical height interpolation |
| Vertintml | ml2pl, ml2hl | Vertical interpolation on hybrid sigma pressure coordinates |
C. Standard name table
The following CF standard names are supported by CDO.
| CF standard name | Units | GRIB 1 code | variable name |
| surface_geopotential | m2 s-2 | 129 | geosp |
| air_temperature | K | 130 | ta |
| specific_humidity | 1 | 133 | hus |
| surface_air_pressure | Pa | 134 | aps |
| air_pressure_at_sea_level | Pa | 151 | psl |
| geopotential_height | m | 156 | zg |
D. Grid description examples
D.1 Example of a curvilinear grid description
Here is an example for the CDO description of a curvilinear grid. xvals/yvals describe the positions of the 6x5 quadrilateral grid cells. The first 4 values of xbounds/ybounds are the corners of the first grid cell.
gridtype = curvilinear gridsize = 30 xsize = 6 ysize = 5 xvals = -21 -11 0 11 21 30 -25 -13 0 13 25 36 -31 -16 0 16 31 43 -38 -21 0 21 38 52 -51 -30 0 30 51 64 xbounds = -23 -14 -17 -28 -14 -5 -6 -17 -5 5 6 -6 5 14 17 6 14 23 28 17 23 32 38 28 -28 -17 -21 -34 -17 -6 -7 -21 -6 6 7 -7 6 17 21 7 17 28 34 21 28 38 44 34 -34 -21 -27 -41 -21 -7 -9 -27 -7 7 9 -9 7 21 27 9 21 34 41 27 34 44 52 41 -41 -27 -35 -51 -27 -9 -13 -35 -9 9 13 -13 9 27 35 13 27 41 51 35 41 52 63 51 -51 -35 -51 -67 -35 -13 -21 -51 -13 13 21 -21 13 35 51 21 35 51 67 51 51 63 77 67 yvals = 29 32 32 32 29 26 39 42 42 42 39 35 48 51 52 51 48 43 57 61 62 61 57 51 65 70 72 70 65 58 ybounds = 23 26 36 32 26 27 37 36 27 27 37 37 27 26 36 37 26 23 32 36 23 19 28 32 32 36 45 41 36 37 47 45 37 37 47 47 37 36 45 47 36 32 41 45 32 28 36 41 41 45 55 50 45 47 57 55 47 47 57 57 47 45 55 57 45 41 50 55 41 36 44 50 50 55 64 58 55 57 67 64 57 57 67 67 57 55 64 67 55 50 58 64 50 44 51 58 58 64 72 64 64 67 77 72 67 67 77 77 67 64 72 77 64 58 64 72 58 51 56 64
| Figure D.1.: | Orthographic and Robinson projection of the curvilinear grid, the first grid cell is colored red |
D.2 Example description for an unstructured grid
Here is an example of the CDO description for an unstructured grid. xvals/yvals describe the positions of 30 independent hexagonal grid cells. The first 6 values of xbounds/ybounds are the corners of the first grid cell. The grid cell corners have to rotate counterclockwise. The first grid cell is colored red.
gridtype = unstructured gridsize = 30 nvertex = 6 xvals = -36 36 0 -18 18 108 72 54 90 180 144 126 162 -108 -144 -162 -126 -72 -90 -54 0 72 36 144 108 -144 180 -72 -108 -36 xbounds = 339 0 0 288 288 309 21 51 72 72 0 0 0 16 21 0 339 344 340 0 -0 344 324 324 20 36 36 16 0 0 93 123 144 144 72 72 72 88 93 72 51 56 52 72 72 56 36 36 92 108 108 88 72 72 165 195 216 216 144 144 144 160 165 144 123 128 124 144 144 128 108 108 164 180 180 160 144 144 237 267 288 288 216 216 216 232 237 216 195 200 196 216 216 200 180 180 236 252 252 232 216 216 288 304 309 288 267 272 268 288 288 272 252 252 308 324 324 304 288 288 345 324 324 36 36 15 36 36 108 108 87 57 20 15 36 57 52 36 108 108 180 180 159 129 92 87 108 129 124 108 180 180 252 252 231 201 164 159 180 201 196 180 252 252 324 324 303 273 236 231 252 273 268 252 308 303 324 345 340 324 yvals = 58 58 32 0 0 58 32 0 0 58 32 0 0 58 32 0 0 32 0 0 -58 -58 -32 -58 -32 -58 -32 -58 -32 -32 ybounds = 41 53 71 71 53 41 41 41 53 71 71 53 11 19 41 53 41 19 -19 -7 11 19 7 -11 -19 -11 7 19 11 -7 41 41 53 71 71 53 11 19 41 53 41 19 -19 -7 11 19 7 -11 -19 -11 7 19 11 -7 41 41 53 71 71 53 11 19 41 53 41 19 -19 -7 11 19 7 -11 -19 -11 7 19 11 -7 41 41 53 71 71 53 11 19 41 53 41 19 -19 -7 11 19 7 -11 -19 -11 7 19 11 -7 11 19 41 53 41 19 -19 -7 11 19 7 -11 -19 -11 7 19 11 -7 -41 -53 -71 -71 -53 -41 -53 -71 -71 -53 -41 -41 -19 -41 -53 -41 -19 -11 -53 -71 -71 -53 -41 -41 -19 -41 -53 -41 -19 -11 -53 -71 -71 -53 -41 -41 -19 -41 -53 -41 -19 -11 -53 -71 -71 -53 -41 -41 -19 -41 -53 -41 -19 -11 -19 -41 -53 -41 -19 -11
Operator catalog
Information
| info | Dataset information listed by parameter identifier |
| infon | Dataset information listed by parameter name |
| cinfo | Compact information listed by parameter name |
| map | Dataset information and simple map |
| sinfo | Short information listed by parameter identifier |
| sinfon | Short information listed by parameter name |
| xsinfo | Extra short information listed by parameter name |
| xsinfop | Extra short information listed by parameter identifier |
| diff | Compare two datasets listed by parameter id |
| diffn | Compare two datasets listed by parameter name |
| npar | Number of parameters |
| nlevel | Number of levels |
| nyear | Number of years |
| nmon | Number of months |
| ndate | Number of dates |
| ntime | Number of timesteps |
| ngridpoints | Number of gridpoints |
| ngrids | Number of horizontal grids |
| showformat | Show file format |
| showcode | Show code numbers |
| showname | Show variable names |
| showstdname | Show standard names |
| showlevel | Show levels |
| showltype | Show GRIB level types |
| showyear | Show years |
| showmon | Show months |
| showdate | Show date information |
| showtime | Show time information |
| showtimestamp | Show timestamp |
| showchunkspec | Show chunk specification |
| showfilter | Show filter specification |
| showattribute | Show a global attribute or a variable attribute |
| partab | Parameter table |
| codetab | Parameter code table |
| griddes | Grid description |
| zaxisdes | Z-axis description |
| vct | Vertical coordinate table |
File operations
| apply | Apply operators on each input file. |
| copy | Copy datasets |
| clone | Clone datasets |
| cat | Concatenate datasets |
| tee | Duplicate a data stream |
| pack | Pack data |
| unpack | Unpack data |
| setchunkspec | Specify chunking |
| setfilter | Specify filter |
| bitrounding | Bit rounding |
| replace | Replace variables |
| duplicate | Duplicates a dataset |
| mergegrid | Merge grid |
| merge | Merge datasets with different fields |
| mergetime | Merge datasets sorted by date and time |
| splitcode | Split code numbers |
| splitparam | Split parameter identifiers |
| splitname | Split variable names |
| splitlevel | Split levels |
| splitgrid | Split grids |
| splitzaxis | Split z-axes |
| splittabnum | Split parameter table numbers |
| splithour | Split hours |
| splitday | Split days |
| splitseas | Split seasons |
| splityear | Split years |
| splityearmon | Split in years and months |
| splitmon | Split months |
| splitsel | Split time selection |
| splitdate | Splits a file into dates |
| distgrid | Distribute horizontal grid |
| collgrid | Collect horizontal grid |
Selection
| select | Select fields |
| delete | Delete fields |
| selmulti | Select multiple fields |
| delmulti | Delete multiple fields |
| changemulti | Change identication of multiple fields |
| selparam | Select parameters by identifier |
| delparam | Delete parameters by identifier |
| selcode | Select parameters by code number |
| delcode | Delete parameters by code number |
| selname | Select parameters by name |
| delname | Delete parameters by name |
| selstdname | Select parameters by standard name |
| sellevel | Select levels |
| sellevidx | Select levels by index |
| selgrid | Select grids |
| selzaxis | Select z-axes |
| selzaxisname | Select z-axes by name |
| selltype | Select GRIB level types |
| seltabnum | Select parameter table numbers |
| seltimestep | Select timesteps |
| seltime | Select times |
| selhour | Select hours |
| selday | Select days |
| selmonth | Select months |
| selyear | Select years |
| selseason | Select seasons |
| seldate | Select dates |
| selsmon | Select single month |
| sellonlatbox | Select a longitude/latitude box |
| selindexbox | Select an index box |
| selregion | Select cells inside regions |
| selcircle | Select cells inside a circle |
| selgridcell | Select grid cells |
| delgridcell | Delete grid cells |
| samplegrid | Resample grid |
| selyearidx | Select year by index |
| seltimeidx | Select timestep by index |
| bottomvalue | Extract bottom level |
| topvalue | Extract top level |
| isosurface | Extract isosurface |
Conditional selection
| ifthen | If then |
| ifnotthen | If not then |
| ifthenelse | If then else |
| ifthenc | If then constant |
| ifnotthenc | If not then constant |
| reducegrid | Reduce input file variables to locations, where mask is non-zero. |
Comparison
| eq | Equal |
| ne | Not equal |
| le | Less equal |
| lt | Less than |
| ge | Greater equal |
| gt | Greater than |
| eqc | Equal constant |
| nec | Not equal constant |
| lec | Less equal constant |
| ltc | Less than constant |
| gec | Greater equal constant |
| gtc | Greater than constant |
| ymoneq | Compare time series with Equal |
| ymonne | Compare time series with NotEqual |
| ymonle | Compare time series with LessEqual |
| ymonlt | Compares if time series with LessThan |
| ymonge | Compares if time series with GreaterEqual |
| ymongt | Compares if time series with GreaterThan |
| yseaseq | Compare time series with Equal |
| yseasne | Compare time series with NotEqual |
| yseasle | Compare time series with LessEqual |
| yseaslt | Compares if time series with LessThan |
| yseasge | Compares if time series with GreaterEqual |
| yseasgt | Compares if time series with GreaterThan |
Modification
| setattribute | Set attributes |
| delattribute | Delete attributes |
| setpartabp | Set parameter table |
| setpartabn | Set parameter table |
| setcodetab | Set parameter code table |
| setcode | Set code number |
| setparam | Set parameter identifier |
| setname | Set variable name |
| setstdname | Set standard name |
| setunit | Set variable unit |
| setlevel | Set level |
| setltype | Set GRIB level type |
| setmaxsteps | Set max timesteps |
| setdate | Set date |
| settime | Set time of the day |
| setday | Set day |
| setmon | Set month |
| setyear | Set year |
| settunits | Set time units |
| settaxis | Set time axis |
| settbounds | Set time bounds |
| setreftime | Set reference time |
| setcalendar | Set calendar |
| shifttime | Shift timesteps |
| chcode | Change code number |
| chparam | Change parameter identifier |
| chname | Change variable or coordinate name |
| chunit | Change variable unit |
| chlevel | Change level |
| chlevelc | Change level of one code |
| chlevelv | Change level of one variable |
| setgrid | Set grid |
| setgridtype | Set grid type |
| setgridarea | Set grid cell area |
| setgridmask | Set grid mask |
| setprojparams | Set proj params |
| setzaxis | Set z-axis |
| genlevelbounds | Generate level bounds |
| invertlat | Invert latitudes |
| invertlev | Invert levels |
| shiftx | Shift x |
| shifty | Shift y |
| maskregion | Mask regions |
| masklonlatbox | Mask a longitude/latitude box |
| maskindexbox | Mask an index box |
| setclonlatbox | Set a longitude/latitude box to constant |
| setcindexbox | Set an index box to constant |
| enlarge | Enlarge fields |
| setmissval | Set a new missing value |
| setctomiss | Set constant to missing value |
| setmisstoc | Set missing value to constant |
| setrtomiss | Set range to missing value |
| setvrange | Set valid range |
| setmisstonn | Set missing value to nearest neighbor |
| setmisstodis | Set missing value to distance-weighted average |
| vertfillmiss | Vertical filling of missing values |
| timfillmiss | Temporal filling of missing values |
| setgridcell | Set the value of a grid cell |
Arithmetic
| expr | Evaluate expressions |
| exprf | Evaluate expressions script |
| aexpr | Evaluate expressions and append results |
| aexprf | Evaluate expression script and append results |
| abs | Absolute value |
| int | Integer value |
| nint | Nearest integer value |
| pow | Power |
| sqr | Square |
| sqrt | Square root |
| exp | Exponential |
| ln | Natural logarithm |
| log10 | Base 10 logarithm |
| sin | Sine |
| cos | Cosine |
| tan | Tangent |
| asin | Arc sine |
| acos | Arc cosine |
| atan | Arc tangent |
| reci | Reciprocal value |
| not | Logical NOT |
| addc | Add a constant |
| subc | Subtract a constant |
| mulc | Multiply with a constant |
| divc | Divide by a constant |
| minc | Minimum of a field and a constant |
| maxc | Maximum of a field and a constant |
| add | Add two fields |
| sub | Subtract two fields |
| mul | Multiply two fields |
| div | Divide two fields |
| min | Minimum of two fields |
| max | Maximum of two fields |
| atan2 | Arc tangent of two fields |
| setmiss | Set missing values |
| dayadd | Add daily time series |
| daysub | Subtract daily time series |
| daymul | Multiply daily time series |
| daydiv | Divide daily time series |
| monadd | Add monthly time series |
| monsub | Subtract monthly time series |
| monmul | Multiply monthly time series |
| mondiv | Divide monthly time series |
| yearadd | Add yearly time series |
| yearsub | Subtract yearly time series |
| yearmul | Multiply yearly time series |
| yeardiv | Divide yearly time series |
| yhouradd | Add multi-year hourly time series |
| yhoursub | Subtract multi-year hourly time series |
| yhourmul | Multiply multi-year hourly time series |
| yhourdiv | Divide multi-year hourly time series |
| ydayadd | Add multi-year daily time series |
| ydaysub | Subtract multi-year daily time series |
| ydaymul | Multiply multi-year daily time series |
| ydaydiv | Divide multi-year daily time series |
| ymonadd | Add multi-year monthly time series |
| ymonsub | Subtract multi-year monthly time series |
| ymonmul | Multiply multi-year monthly time series |
| ymondiv | Divide multi-year monthly time series |
| yseasadd | Add multi-year seasonal time series |
| yseassub | Subtract multi-year seasonal time series |
| yseasmul | Multiply multi-year seasonal time series |
| yseasdiv | Divide multi-year seasonal time series |
| muldpm | Multiply with days per month |
| divdpm | Divide by days per month |
| muldpy | Multiply with days per year |
| divdpy | Divide by days per year |
| mulcoslat | Multiply with the cosine of the latitude |
| divcoslat | Divide by cosine of the latitude |
Statistical values
| timcumsum | Cumulative sum over all timesteps |
| consecsum | Consecutive Sum |
| consects | Consecutive Timesteps |
| varsmin | Variables minimum |
| varsmax | Variables maximum |
| varsrange | Variables range |
| varssum | Variables sum |
| varsmean | Variables mean |
| varsavg | Variables average |
| varsstd | Variables standard deviation |
| varsstd1 | Variables standard deviation (n-1) |
| varsvar | Variables variance |
| varsvar1 | Variables variance (n-1) |
| ensmin | Ensemble minimum |
| ensmax | Ensemble maximum |
| ensrange | Ensemble range |
| enssum | Ensemble sum |
| ensmean | Ensemble mean |
| ensavg | Ensemble average |
| ensstd | Ensemble standard deviation |
| ensstd1 | Ensemble standard deviation (n-1) |
| ensvar | Ensemble variance |
| ensvar1 | Ensemble variance (n-1) |
| ensskew | Ensemble skewness |
| enskurt | Ensemble kurtosis |
| ensmedian | Ensemble median |
| enspctl | Ensemble percentiles |
| ensrkhistspace | Ranked Histogram averaged over space |
| ensrkhisttime | Ranked Histogram averaged over time |
| ensroc | Ensemble Receiver Operating characteristics |
| enscrps | Ensemble CRPS and decomposition |
| ensbrs | Ensemble Brier score |
| fldmin | Field minimum |
| fldmax | Field maximum |
| fldrange | Field range |
| fldsum | Field sum |
| fldint | Field integral |
| fldmean | Field mean |
| fldavg | Field average |
| fldstd | Field standard deviation |
| fldstd1 | Field standard deviation (n-1) |
| fldvar | Field variance |
| fldvar1 | Field variance (n-1) |
| fldskew | Field skewness |
| fldkurt | Field kurtosis |
| fldmedian | Field median |
| fldcount | Field count |
| fldpctl | Field percentiles |
| zonmin | Zonal minimum |
| zonmax | Zonal maximum |
| zonrange | Zonal range |
| zonsum | Zonal sum |
| zonmean | Zonal mean |
| zonavg | Zonal average |
| zonstd | Zonal standard deviation |
| zonstd1 | Zonal standard deviation (n-1) |
| zonvar | Zonal variance |
| zonvar1 | Zonal variance (n-1) |
| zonskew | Zonal skewness |
| zonkurt | Zonal kurtosis |
| zonmedian | Zonal median |
| zonpctl | Zonal percentiles |
| mermin | Meridional minimum |
| mermax | Meridional maximum |
| merrange | Meridional range |
| mersum | Meridional sum |
| mermean | Meridional mean |
| meravg | Meridional average |
| merstd | Meridional standard deviation |
| merstd1 | Meridional standard deviation (n-1) |
| mervar | Meridional variance |
| mervar1 | Meridional variance (n-1) |
| merskew | Meridional skewness |
| merkurt | Meridional kurtosis |
| mermedian | Meridional median |
| merpctl | Meridional percentiles |
| gridboxmin | Gridbox minimum |
| gridboxmax | Gridbox maximum |
| gridboxrange | Gridbox range |
| gridboxsum | Gridbox sum |
| gridboxmean | Gridbox mean |
| gridboxavg | Gridbox average |
| gridboxstd | Gridbox standard deviation |
| gridboxstd1 | Gridbox standard deviation (n-1) |
| gridboxvar | Gridbox variance |
| gridboxvar1 | Gridbox variance (n-1) |
| gridboxskew | Gridbox skewness |
| gridboxkurt | Gridbox kurtosis |
| gridboxmedian | Gridbox median |
| remapmin | Remap minimum |
| remapmax | Remap maximum |
| remaprange | Remap range |
| remapsum | Remap sum |
| remapmean | Remap mean |
| remapavg | Remap average |
| remapstd | Remap standard deviation |
| remapstd1 | Remap standard deviation (n-1) |
| remapvar | Remap variance |
| remapvar1 | Remap variance (n-1) |
| remapskew | Remap skewness |
| remapkurt | Remap kurtosis |
| remapmedian | Remap median |
| vertmin | Vertical minimum |
| vertmax | Vertical maximum |
| vertrange | Vertical range |
| vertsum | Vertical sum |
| vertmean | Vertical mean |
| vertavg | Vertical average |
| vertstd | Vertical standard deviation |
| vertstd1 | Vertical standard deviation (n-1) |
| vertvar | Vertical variance |
| vertvar1 | Vertical variance (n-1) |
| timselmin | Time selection minimum |
| timselmax | Time selection maximum |
| timselrange | Time selection range |
| timselsum | Time selection sum |
| timselmean | Time selection mean |
| timselavg | Time selection average |
| timselstd | Time selection standard deviation |
| timselstd1 | Time selection standard deviation (n-1) |
| timselvar | Time selection variance |
| timselvar1 | Time selection variance (n-1) |
| timselpctl | Time range percentiles |
| runmin | Running minimum |
| runmax | Running maximum |
| runrange | Running range |
| runsum | Running sum |
| runmean | Running mean |
| runavg | Running average |
| runstd | Running standard deviation |
| runstd1 | Running standard deviation (n-1) |
| runvar | Running variance |
| runvar1 | Running variance (n-1) |
| runpctl | Running percentiles |
| timmin | Time minimum |
| timmax | Time maximum |
| timminidx | Index of time minimum |
| timmaxidx | Index of time maximum |
| timrange | Time range |
| timsum | Time sum |
| timmean | Time mean |
| timavg | Time average |
| timstd | Time standard deviation |
| timstd1 | Time standard deviation (n-1) |
| timvar | Time variance |
| timvar1 | Time variance (n-1) |
| timpctl | Time percentiles |
| hourmin | Hourly minimum |
| hourmax | Hourly maximum |
| hourrange | Hourly range |
| hoursum | Hourly sum |
| hourmean | Hourly mean |
| houravg | Hourly average |
| hourstd | Hourly standard deviation |
| hourstd1 | Hourly standard deviation (n-1) |
| hourvar | Hourly variance |
| hourvar1 | Hourly variance (n-1) |
| hourpctl | Hourly percentiles |
| daymin | Daily minimum |
| daymax | Daily maximum |
| dayrange | Daily range |
| daysum | Daily sum |
| daymean | Daily mean |
| dayavg | Daily average |
| daystd | Daily standard deviation |
| daystd1 | Daily standard deviation (n-1) |
| dayvar | Daily variance |
| dayvar1 | Daily variance (n-1) |
| daypctl | Daily percentiles |
| monmin | Monthly minimum |
| monmax | Monthly maximum |
| monrange | Monthly range |
| monsum | Monthly sum |
| monmean | Monthly mean |
| monavg | Monthly average |
| monstd | Monthly standard deviation |
| monstd1 | Monthly standard deviation (n-1) |
| monvar | Monthly variance |
| monvar1 | Monthly variance (n-1) |
| monpctl | Monthly percentiles |
| yearmonmean | Yearly mean from monthly data |
| yearmin | Yearly minimum |
| yearmax | Yearly maximum |
| yearminidx | Index of yearly minimum |
| yearmaxidx | Index of yearly maximum |
| yearrange | Yearly range |
| yearsum | Yearly sum |
| yearmean | Yearly mean |
| yearavg | Yearly average |
| yearstd | Yearly standard deviation |
| yearstd1 | Yearly standard deviation (n-1) |
| yearvar | Yearly variance |
| yearvar1 | Yearly variance (n-1) |
| yearpctl | Yearly percentiles |
| seasmin | Seasonal minimum |
| seasmax | Seasonal maximum |
| seasrange | Seasonal range |
| seassum | Seasonal sum |
| seasmean | Seasonal mean |
| seasavg | Seasonal average |
| seasstd | Seasonal standard deviation |
| seasstd1 | Seasonal standard deviation (n-1) |
| seasvar | Seasonal variance |
| seasvar1 | Seasonal variance (n-1) |
| seaspctl | Seasonal percentiles |
| yhourmin | Multi-year hourly minimum |
| yhourmax | Multi-year hourly maximum |
| yhourrange | Multi-year hourly range |
| yhoursum | Multi-year hourly sum |
| yhourmean | Multi-year hourly mean |
| yhouravg | Multi-year hourly average |
| yhourstd | Multi-year hourly standard deviation |
| yhourstd1 | Multi-year hourly standard deviation (n-1) |
| yhourvar | Multi-year hourly variance |
| yhourvar1 | Multi-year hourly variance (n-1) |
| dhourmin | Multi-day hourly minimum |
| dhourmax | Multi-day hourly maximum |
| dhourrange | Multi-day hourly range |
| dhoursum | Multi-day hourly sum |
| dhourmean | Multi-day hourly mean |
| dhouravg | Multi-day hourly average |
| dhourstd | Multi-day hourly standard deviation |
| dhourstd1 | Multi-day hourly standard deviation (n-1) |
| dhourvar | Multi-day hourly variance |
| dhourvar1 | Multi-day hourly variance (n-1) |
| dminutemin | Multi-day by the minute minimum |
| dminutemax | Multi-day by the minute maximum |
| dminuterange | Multi-day by the minute range |
| dminutesum | Multi-day by the minute sum |
| dminutemean | Multi-day by the minute mean |
| dminuteavg | Multi-day by the minute average |
| dminutestd | Multi-day by the minute standard deviation |
| dminutestd1 | Multi-day by the minute standard deviation (n-1) |
| dminutevar | Multi-day by the minute variance |
| dminutevar1 | Multi-day by the minute variance (n-1) |
| ydaymin | Multi-year daily minimum |
| ydaymax | Multi-year daily maximum |
| ydayrange | Multi-year daily range |
| ydaysum | Multi-year daily sum |
| ydaymean | Multi-year daily mean |
| ydayavg | Multi-year daily average |
| ydaystd | Multi-year daily standard deviation |
| ydaystd1 | Multi-year daily standard deviation (n-1) |
| ydayvar | Multi-year daily variance |
| ydayvar1 | Multi-year daily variance (n-1) |
| ydaypctl | Multi-year daily percentiles |
| ymonmin | Multi-year monthly minimum |
| ymonmax | Multi-year monthly maximum |
| ymonrange | Multi-year monthly range |
| ymonsum | Multi-year monthly sum |
| ymonmean | Multi-year monthly mean |
| ymonavg | Multi-year monthly average |
| ymonstd | Multi-year monthly standard deviation |
| ymonstd1 | Multi-year monthly standard deviation (n-1) |
| ymonvar | Multi-year monthly variance |
| ymonvar1 | Multi-year monthly variance (n-1) |
| ymonpctl | Multi-year monthly percentiles |
| yseasmin | Multi-year seasonal minimum |
| yseasmax | Multi-year seasonal maximum |
| yseasrange | Multi-year seasonal range |
| yseassum | Multi-year seasonal sum |
| yseasmean | Multi-year seasonal mean |
| yseasavg | Multi-year seasonal average |
| yseasstd | Multi-year seasonal standard deviation |
| yseasstd1 | Multi-year seasonal standard deviation (n-1) |
| yseasvar | Multi-year seasonal variance |
| yseasvar1 | Multi-year seasonal variance (n-1) |
| yseaspctl | Multi-year seasonal percentiles |
| ydrunmin | Multi-year daily running minimum |
| ydrunmax | Multi-year daily running maximum |
| ydrunsum | Multi-year daily running sum |
| ydrunmean | Multi-year daily running mean |
| ydrunavg | Multi-year daily running average |
| ydrunstd | Multi-year daily running standard deviation |
| ydrunstd1 | Multi-year daily running standard deviation (n-1) |
| ydrunvar | Multi-year daily running variance |
| ydrunvar1 | Multi-year daily running variance (n-1) |
| ydrunpctl | Multi-year daily running percentiles |
Correlation and co.
| fldcor | Correlation in grid space |
| timcor | Correlation over time |
| fldcovar | Covariance in grid space |
| timcovar | Covariance over time |
Regression
| regres | Regression |
| detrend | Detrend |
| trend | Trend |
| addtrend | Add trend |
| subtrend | Subtract trend |
Eof
| eof | Calculate EOFs in spatial or time space |
| eoftime | Calculate EOFs in time space |
| eofspatial | Calculate EOFs in spatial space |
| eof3d | Calculate 3-Dimensional EOFs in time space |
| eofcoeff | Calculate principal coefficients of EOFs |
Interpolation
| remapbil | Bilinear interpolation |
| genbil | Generate bilinear interpolation weights |
| remapbic | Bicubic interpolation |
| genbic | Generate bicubic interpolation weights |
| remapnn | Nearest neighbor remapping |
| gennn | Generate nearest neighbor remap weights |
| remapdis | Distance weighted average remapping |
| gendis | Generate distance weighted average remap weights |
| remapcon | First order conservative remapping |
| gencon | Generate 1st order conservative remap weights |
| remaplaf | Largest area fraction remapping |
| genlaf | Generate largest area fraction remap weights |
| remap | Grid remapping |
| remapeta | Remap vertical hybrid level |
| ml2pl | Model to pressure level interpolation |
| ml2hl | Model to height level interpolation |
| ap2pl | Air pressure to pressure level interpolation |
| gh2hl | Geometric height to height level interpolation |
| intlevel | Linear level interpolation |
| intlevel3d | Linear level interpolation onto a 3D vertical coordinate |
| intlevelx3d | like intlevel3d but with extrapolation |
| inttime | Interpolation between timesteps |
| intntime | Interpolation between timesteps |
| intyear | Interpolation between two years |
Transformation
| sp2gp | Spectral to gridpoint |
| gp2sp | Gridpoint to spectral |
| sp2sp | Spectral to spectral |
| dv2ps | D and V to velocity potential and stream function |
| dv2uv | Divergence and vorticity to U and V wind |
| uv2dv | U and V wind to divergence and vorticity |
| fourier | Fourier transformation |
Import/Export
| import_binary | Import binary data sets |
| import_cmsaf | Import CM-SAF HDF5 files |
| input | ASCII input |
| inputsrv | SERVICE ASCII input |
| inputext | EXTRA ASCII input |
| output | ASCII output |
| outputf | Formatted output |
| outputint | Integer output |
| outputsrv | SERVICE ASCII output |
| outputext | EXTRA ASCII output |
| outputtab | Table output |
| gmtxyz | GMT xyz format |
| gmtcells | GMT multiple segment format |
Miscellaneous
| gradsdes | GrADS data descriptor file |
| after | ECHAM standard post processor |
| bandpass | Bandpass filtering |
| lowpass | Lowpass filtering |
| highpass | Highpass filtering |
| gridarea | Grid cell area |
| gridweights | Grid cell weights |
| smooth | Smooth grid points |
| smooth9 | 9 point smoothing |
| setvals | Set list of old values to new values |
| setrtoc | Set range to constant |
| setrtoc2 | Set range to constant others to constant2 |
| gridcellindex | Get grid cell index from lon/lat point |
| const | Create a constant field |
| random | Create a field with random numbers |
| topo | Create a field with topography |
| seq | Create a time series |
| stdatm | Create values for pressure and temperature for hydrostatic atmosphere |
| timsort | Sort over the time |
| uvDestag | Destaggering of u/v wind components |
| rotuvNorth | Rotate u/v wind to North pole. |
| projuvLatLon | Cylindrical Equidistant projection |
| rotuvb | Backward rotation |
| mrotuvb | Backward rotation of MPIOM data |
| mastrfu | Mass stream function |
| pressure_half | Pressure on half-levels |
| pressure | Pressure on full-levels |
| delta_pressure | Pressure difference of half-levels |
| sealevelpressure | Sea level pressure |
| gheight | Geopotential height on full-levels |
| gheight_half | Geopotential height on half-levels |
| air_density | Air density |
| adisit | Potential temperature to in-situ temperature |
| adipot | In-situ temperature to potential temperature |
| rhopot | Calculates potential density |
| histcount | Histogram count |
| histsum | Histogram sum |
| histmean | Histogram mean |
| histfreq | Histogram frequency |
| sethalo | Set the bounds of a field |
| wct | Windchill temperature |
| fdns | Frost days where no snow index per time period |
| strwin | Strong wind days index per time period |
| strbre | Strong breeze days index per time period |
| strgal | Strong gale days index per time period |
| hurr | Hurricane days index per time period |
| cmorlite | CMOR lite |
| verifygrid | Verify grid coordinates |
| hpdegrade | Degrade healpix |
| hpupgrade | Upgrade healpix |
NCL
| uv2vr_cfd | U and V wind to relative vorticity |
| uv2dv_cfd | U and V wind to divergence |
Cmor
| cmor | Climate Model Output Rewriting |
Magics
| contour | Contour plot |
| shaded | Shaded contour plot |
| grfill | Shaded gridfill plot |
| vector | Vector arrows plot |
| graph | Line graph plot |
Climate indices
| eca_cdd | Consecutive dry days index per time period |
| etccdi_cdd | Consecutive dry days index per time period |
| eca_cfd | Consecutive frost days index per time period |
| eca_csu | Consecutive summer days index per time period |
| eca_cwd | Consecutive wet days index per time period |
| etccdi_cwd | Consecutive wet days index per time period |
| eca_cwdi | Cold wave duration index wrt mean of reference period |
| eca_cwfi | Cold-spell days index wrt 10th percentile of reference period |
| etccdi_csdi | Cold-spell duration index |
| eca_etr | Intra-period extreme temperature range |
| eca_fd | Frost days index per time period |
| etccdi_fd | Frost days index per time period |
| eca_gsl | Growing season length index |
| eca_hd | Heating degree days per time period |
| eca_hwdi | Heat wave duration index wrt mean of reference period |
| eca_hwfi | Warm spell days index wrt 90th percentile of reference period |
| etccdi_wsdi | Warm Spell Duration Index |
| eca_id | Ice days index per time period |
| etccdi_id | Ice days index per time period |
| eca_r75p | Moderate wet days wrt 75th percentile of reference period |
| eca_r75ptot | Precipitation percent due to R75p days |
| eca_r90p | Wet days wrt 90th percentile of reference period |
| eca_r90ptot | Precipitation percent due to R90p days |
| eca_r95p | Very wet days wrt 95th percentile of reference period |
| eca_r95ptot | Precipitation percent due to R95p days |
| eca_r99p | Extremely wet days wrt 99th percentile of reference period |
| eca_r99ptot | Precipitation percent due to R99p days |
| eca_pd | Precipitation days index per time period |
| eca_r10mm | Heavy precipitation days index per time period |
| eca_r20mm | Very heavy precipitation days index per time period |
| etccdi_r1mm | Precipitation days index per time period |
| eca_rr1 | Wet days index per time period |
| eca_rx1day | Highest one day precipitation amount per time period |
| etccdi_rx1day | Maximum 1-day Precipitation |
| eca_rx5day | Highest five-day precipitation amount per time period |
| etccdi_rx5day | Highest five-day precipitation amount per time period |
| eca_sdii | Simple daily intensity index per time period |
| eca_su | Summer days index per time period |
| etccdi_su | Summer days index per time period |
| eca_tg10p | Cold days percent wrt 10th percentile of reference period |
| eca_tg90p | Warm days percent wrt 90th percentile of reference period |
| eca_tn10p | Cold nights percent wrt 10th percentile of reference period |
| eca_tn90p | Warm nights percent wrt 90th percentile of reference period |
| eca_tr | Tropical nights index per time period |
| etccdi_tr | Tropical nights index per time period |
| eca_tx10p | Very cold days percent wrt 10th percentile of reference period |
| eca_tx90p | Very warm days percent wrt 90th percentile of reference period |
| etccdi_tx90p | Percentage of Days when Daily Maximum Temperature is Above the 90th Percentile |
| etccdi_tx10p | Percentage of Days when Daily Maximum Temperature is Below the 10th Percentile |
| etccdi_tn90p | Percentage of Days when Daily Minimum Temperature is Above the 90th Percentile |
| etccdi_tn10p | Percentage of Days when Daily Minimum Temperature is Below the 10th Percentile |
| etccdi_r95p | Annual Total Precipitation when Daily Precipitation Exceeds the 95th Percentile of Wet Day Precipitation |
| etccdi_r99p | Annual Total Precipitation when Daily Precipitation Exceeds the 99th Percentile of Wet Day Precipitation |
Alphabetic List of Operators
| abs | Absolute value |
| acos | Arc cosine |
| addc | Add a constant |
| addtrend | Add trend |
| add | Add two fields |
| adipot | In-situ temperature to potential temperature |
| adisit | Potential temperature to in-situ temperature |
| aexprf | Evaluate expression script and append results |
| aexpr | Evaluate expressions and append results |
| after | ECHAM standard post processor |
| air_density | Air density |
| ap2pl | Air pressure to pressure level interpolation |
| apply | Apply operators on each input file. |
| asin | Arc sine |
| atan2 | Arc tangent of two fields |
| atan | Arc tangent |
| bandpass | Bandpass filtering |
| bitrounding | Bit rounding |
| bottomvalue | Extract bottom level |
| cat | Concatenate datasets |
| changemulti | Change identication of multiple fields |
| chcode | Change code number |
| chlevelc | Change level of one code |
| chlevelv | Change level of one variable |
| chlevel | Change level |
| chname | Change variable or coordinate name |
| chparam | Change parameter identifier |
| chunit | Change variable unit |
| cinfo | Compact information listed by parameter name |
| clone | Clone datasets |
| cmorlite | CMOR lite |
| cmor | Climate Model Output Rewriting |
| codetab | Parameter code table |
| collgrid | Collect horizontal grid |
| consecsum | Consecutive Sum |
| consects | Consecutive Timesteps |
| const | Create a constant field |
| contour | Contour plot |
| copy | Copy datasets |
| cos | Cosine |
| dayadd | Add daily time series |
| dayavg | Daily average |
| daydiv | Divide daily time series |
| daymax | Daily maximum |
| daymean | Daily mean |
| daymin | Daily minimum |
| daymul | Multiply daily time series |
| daypctl | Daily percentiles |
| dayrange | Daily range |
| daystd1 | Daily standard deviation (n-1) |
| daystd | Daily standard deviation |
| daysub | Subtract daily time series |
| daysum | Daily sum |
| dayvar1 | Daily variance (n-1) |
| dayvar | Daily variance |
| delattribute | Delete attributes |
| delcode | Delete parameters by code number |
| delete | Delete fields |
| delgridcell | Delete grid cells |
| delmulti | Delete multiple fields |
| delname | Delete parameters by name |
| delparam | Delete parameters by identifier |
| delta_pressure | Pressure difference of half-levels |
| detrend | Detrend |
| dhouravg | Multi-day hourly average |
| dhourmax | Multi-day hourly maximum |
| dhourmean | Multi-day hourly mean |
| dhourmin | Multi-day hourly minimum |
| dhourrange | Multi-day hourly range |
| dhourstd1 | Multi-day hourly standard deviation (n-1) |
| dhourstd | Multi-day hourly standard deviation |
| dhoursum | Multi-day hourly sum |
| dhourvar1 | Multi-day hourly variance (n-1) |
| dhourvar | Multi-day hourly variance |
| diffn | Compare two datasets listed by parameter name |
| diff | Compare two datasets listed by parameter id |
| distgrid | Distribute horizontal grid |
| divcoslat | Divide by cosine of the latitude |
| divc | Divide by a constant |
| divdpm | Divide by days per month |
| divdpy | Divide by days per year |
| div | Divide two fields |
| dminuteavg | Multi-day by the minute average |
| dminutemax | Multi-day by the minute maximum |
| dminutemean | Multi-day by the minute mean |
| dminutemin | Multi-day by the minute minimum |
| dminuterange | Multi-day by the minute range |
| dminutestd1 | Multi-day by the minute standard deviation (n-1) |
| dminutestd | Multi-day by the minute standard deviation |
| dminutesum | Multi-day by the minute sum |
| dminutevar1 | Multi-day by the minute variance (n-1) |
| dminutevar | Multi-day by the minute variance |
| duplicate | Duplicates a dataset |
| dv2ps | D and V to velocity potential and stream function |
| dv2uv | Divergence and vorticity to U and V wind |
| eca_cdd | Consecutive dry days index per time period |
| eca_cfd | Consecutive frost days index per time period |
| eca_csu | Consecutive summer days index per time period |
| eca_cwdi | Cold wave duration index wrt mean of reference period |
| eca_cwd | Consecutive wet days index per time period |
| eca_cwfi | Cold-spell days index wrt 10th percentile of reference period |
| eca_etr | Intra-period extreme temperature range |
| eca_fd | Frost days index per time period |
| eca_gsl | Growing season length index |
| eca_hd | Heating degree days per time period |
| eca_hwdi | Heat wave duration index wrt mean of reference period |
| eca_hwfi | Warm spell days index wrt 90th percentile of reference period |
| eca_id | Ice days index per time period |
| eca_pd | Precipitation days index per time period |
| eca_r10mm | Heavy precipitation days index per time period |
| eca_r20mm | Very heavy precipitation days index per time period |
| eca_r75ptot | Precipitation percent due to R75p days |
| eca_r75p | Moderate wet days wrt 75th percentile of reference period |
| eca_r90ptot | Precipitation percent due to R90p days |
| eca_r90p | Wet days wrt 90th percentile of reference period |
| eca_r95ptot | Precipitation percent due to R95p days |
| eca_r95p | Very wet days wrt 95th percentile of reference period |
| eca_r99ptot | Precipitation percent due to R99p days |
| eca_r99p | Extremely wet days wrt 99th percentile of reference period |
| eca_rr1 | Wet days index per time period |
| eca_rx1day | Highest one day precipitation amount per time period |
| eca_rx5day | Highest five-day precipitation amount per time period |
| eca_sdii | Simple daily intensity index per time period |
| eca_su | Summer days index per time period |
| eca_tg10p | Cold days percent wrt 10th percentile of reference period |
| eca_tg90p | Warm days percent wrt 90th percentile of reference period |
| eca_tn10p | Cold nights percent wrt 10th percentile of reference period |
| eca_tn90p | Warm nights percent wrt 90th percentile of reference period |
| eca_tr | Tropical nights index per time period |
| eca_tx10p | Very cold days percent wrt 10th percentile of reference period |
| eca_tx90p | Very warm days percent wrt 90th percentile of reference period |
| enlarge | Enlarge fields |
| ensavg | Ensemble average |
| ensbrs | Ensemble Brier score |
| enscrps | Ensemble CRPS and decomposition |
| enskurt | Ensemble kurtosis |
| ensmax | Ensemble maximum |
| ensmean | Ensemble mean |
| ensmedian | Ensemble median |
| ensmin | Ensemble minimum |
| enspctl | Ensemble percentiles |
| ensrange | Ensemble range |
| ensrkhistspace | Ranked Histogram averaged over space |
| ensrkhisttime | Ranked Histogram averaged over time |
| ensroc | Ensemble Receiver Operating characteristics |
| ensskew | Ensemble skewness |
| ensstd1 | Ensemble standard deviation (n-1) |
| ensstd | Ensemble standard deviation |
| enssum | Ensemble sum |
| ensvar1 | Ensemble variance (n-1) |
| ensvar | Ensemble variance |
| eof3d | Calculate 3-Dimensional EOFs in time space |
| eofcoeff | Calculate principal coefficients of EOFs |
| eofspatial | Calculate EOFs in spatial space |
| eoftime | Calculate EOFs in time space |
| eof | Calculate EOFs in spatial or time space |
| eqc | Equal constant |
| eq | Equal |
| etccdi_cdd | Consecutive dry days index per time period |
| etccdi_csdi | Cold-spell duration index |
| etccdi_cwd | Consecutive wet days index per time period |
| etccdi_fd | Frost days index per time period |
| etccdi_id | Ice days index per time period |
| etccdi_r1mm | Precipitation days index per time period |
| etccdi_r95p | Annual Total Precipitation when Daily Precipitation Exceeds the 95th Percentile of Wet Day Precipitation |
| etccdi_r99p | Annual Total Precipitation when Daily Precipitation Exceeds the 99th Percentile of Wet Day Precipitation |
| etccdi_rx1day | Maximum 1-day Precipitation |
| etccdi_rx5day | Highest five-day precipitation amount per time period |
| etccdi_su | Summer days index per time period |
| etccdi_tn10p | Percentage of Days when Daily Minimum Temperature is Below the 10th Percentile |
| etccdi_tn90p | Percentage of Days when Daily Minimum Temperature is Above the 90th Percentile |
| etccdi_tr | Tropical nights index per time period |
| etccdi_tx10p | Percentage of Days when Daily Maximum Temperature is Below the 10th Percentile |
| etccdi_tx90p | Percentage of Days when Daily Maximum Temperature is Above the 90th Percentile |
| etccdi_wsdi | Warm Spell Duration Index |
| exprf | Evaluate expressions script |
| expr | Evaluate expressions |
| exp | Exponential |
| fdns | Frost days where no snow index per time period |
| fldavg | Field average |
| fldcor | Correlation in grid space |
| fldcount | Field count |
| fldcovar | Covariance in grid space |
| fldint | Field integral |
| fldkurt | Field kurtosis |
| fldmax | Field maximum |
| fldmean | Field mean |
| fldmedian | Field median |
| fldmin | Field minimum |
| fldpctl | Field percentiles |
| fldrange | Field range |
| fldskew | Field skewness |
| fldstd1 | Field standard deviation (n-1) |
| fldstd | Field standard deviation |
| fldsum | Field sum |
| fldvar1 | Field variance (n-1) |
| fldvar | Field variance |
| fourier | Fourier transformation |
| gec | Greater equal constant |
| genbic | Generate bicubic interpolation weights |
| genbil | Generate bilinear interpolation weights |
| gencon | Generate 1st order conservative remap weights |
| gendis | Generate distance weighted average remap weights |
| genlaf | Generate largest area fraction remap weights |
| genlevelbounds | Generate level bounds |
| gennn | Generate nearest neighbor remap weights |
| ge | Greater equal |
| gh2hl | Geometric height to height level interpolation |
| gheight_half | Geopotential height on half-levels |
| gheight | Geopotential height on full-levels |
| gmtcells | GMT multiple segment format |
| gmtxyz | GMT xyz format |
| gp2sp | Gridpoint to spectral |
| gradsdes | GrADS data descriptor file |
| graph | Line graph plot |
| grfill | Shaded gridfill plot |
| gridarea | Grid cell area |
| gridboxavg | Gridbox average |
| gridboxkurt | Gridbox kurtosis |
| gridboxmax | Gridbox maximum |
| gridboxmean | Gridbox mean |
| gridboxmedian | Gridbox median |
| gridboxmin | Gridbox minimum |
| gridboxrange | Gridbox range |
| gridboxskew | Gridbox skewness |
| gridboxstd1 | Gridbox standard deviation (n-1) |
| gridboxstd | Gridbox standard deviation |
| gridboxsum | Gridbox sum |
| gridboxvar1 | Gridbox variance (n-1) |
| gridboxvar | Gridbox variance |
| gridcellindex | Get grid cell index from lon/lat point |
| griddes | Grid description |
| gridweights | Grid cell weights |
| gtc | Greater than constant |
| gt | Greater than |
| highpass | Highpass filtering |
| histcount | Histogram count |
| histfreq | Histogram frequency |
| histmean | Histogram mean |
| histsum | Histogram sum |
| houravg | Hourly average |
| hourmax | Hourly maximum |
| hourmean | Hourly mean |
| hourmin | Hourly minimum |
| hourpctl | Hourly percentiles |
| hourrange | Hourly range |
| hourstd1 | Hourly standard deviation (n-1) |
| hourstd | Hourly standard deviation |
| hoursum | Hourly sum |
| hourvar1 | Hourly variance (n-1) |
| hourvar | Hourly variance |
| hpdegrade | Degrade healpix |
| hpupgrade | Upgrade healpix |
| hurr | Hurricane days index per time period |
| ifnotthenc | If not then constant |
| ifnotthen | If not then |
| ifthenc | If then constant |
| ifthenelse | If then else |
| ifthen | If then |
| import_binary | Import binary data sets |
| import_cmsaf | Import CM-SAF HDF5 files |
| infon | Dataset information listed by parameter name |
| info | Dataset information listed by parameter identifier |
| inputext | EXTRA ASCII input |
| inputsrv | SERVICE ASCII input |
| input | ASCII input |
| intlevel3d | Linear level interpolation onto a 3D vertical coordinate |
| intlevelx3d | like intlevel3d but with extrapolation |
| intlevel | Linear level interpolation |
| intntime | Interpolation between timesteps |
| inttime | Interpolation between timesteps |
| intyear | Interpolation between two years |
| int | Integer value |
| invertlat | Invert latitudes |
| invertlev | Invert levels |
| isosurface | Extract isosurface |
| lec | Less equal constant |
| le | Less equal |
| ln | Natural logarithm |
| log10 | Base 10 logarithm |
| lowpass | Lowpass filtering |
| ltc | Less than constant |
| lt | Less than |
| map | Dataset information and simple map |
| maskindexbox | Mask an index box |
| masklonlatbox | Mask a longitude/latitude box |
| maskregion | Mask regions |
| mastrfu | Mass stream function |
| maxc | Maximum of a field and a constant |
| max | Maximum of two fields |
| meravg | Meridional average |
| mergegrid | Merge grid |
| mergetime | Merge datasets sorted by date and time |
| merge | Merge datasets with different fields |
| merkurt | Meridional kurtosis |
| mermax | Meridional maximum |
| mermean | Meridional mean |
| mermedian | Meridional median |
| mermin | Meridional minimum |
| merpctl | Meridional percentiles |
| merrange | Meridional range |
| merskew | Meridional skewness |
| merstd1 | Meridional standard deviation (n-1) |
| merstd | Meridional standard deviation |
| mersum | Meridional sum |
| mervar1 | Meridional variance (n-1) |
| mervar | Meridional variance |
| minc | Minimum of a field and a constant |
| min | Minimum of two fields |
| ml2hl | Model to height level interpolation |
| ml2pl | Model to pressure level interpolation |
| monadd | Add monthly time series |
| monavg | Monthly average |
| mondiv | Divide monthly time series |
| monmax | Monthly maximum |
| monmean | Monthly mean |
| monmin | Monthly minimum |
| monmul | Multiply monthly time series |
| monpctl | Monthly percentiles |
| monrange | Monthly range |
| monstd1 | Monthly standard deviation (n-1) |
| monstd | Monthly standard deviation |
| monsub | Subtract monthly time series |
| monsum | Monthly sum |
| monvar1 | Monthly variance (n-1) |
| monvar | Monthly variance |
| mrotuvb | Backward rotation of MPIOM data |
| mulcoslat | Multiply with the cosine of the latitude |
| mulc | Multiply with a constant |
| muldpm | Multiply with days per month |
| muldpy | Multiply with days per year |
| mul | Multiply two fields |
| ndate | Number of dates |
| nec | Not equal constant |
| ne | Not equal |
| ngridpoints | Number of gridpoints |
| ngrids | Number of horizontal grids |
| nint | Nearest integer value |
| nlevel | Number of levels |
| nmon | Number of months |
| not | Logical NOT |
| npar | Number of parameters |
| ntime | Number of timesteps |
| nyear | Number of years |
| outputext | EXTRA ASCII output |
| outputf | Formatted output |
| outputint | Integer output |
| outputsrv | SERVICE ASCII output |
| outputtab | Table output |
| output | ASCII output |
| pack | Pack data |
| partab | Parameter table |
| pow | Power |
| pressure_half | Pressure on half-levels |
| pressure | Pressure on full-levels |
| projuvLatLon | Cylindrical Equidistant projection |
| random | Create a field with random numbers |
| reci | Reciprocal value |
| reducegrid | Reduce input file variables to locations, where mask is non-zero. |
| regres | Regression |
| remapavg | Remap average |
| remapbic | Bicubic interpolation |
| remapbil | Bilinear interpolation |
| remapcon | First order conservative remapping |
| remapdis | Distance weighted average remapping |
| remapeta | Remap vertical hybrid level |
| remapkurt | Remap kurtosis |
| remaplaf | Largest area fraction remapping |
| remapmax | Remap maximum |
| remapmean | Remap mean |
| remapmedian | Remap median |
| remapmin | Remap minimum |
| remapnn | Nearest neighbor remapping |
| remaprange | Remap range |
| remapskew | Remap skewness |
| remapstd1 | Remap standard deviation (n-1) |
| remapstd | Remap standard deviation |
| remapsum | Remap sum |
| remapvar1 | Remap variance (n-1) |
| remapvar | Remap variance |
| remap | Grid remapping |
| replace | Replace variables |
| rhopot | Calculates potential density |
| rotuvNorth | Rotate u/v wind to North pole. |
| rotuvb | Backward rotation |
| runavg | Running average |
| runmax | Running maximum |
| runmean | Running mean |
| runmin | Running minimum |
| runpctl | Running percentiles |
| runrange | Running range |
| runstd1 | Running standard deviation (n-1) |
| runstd | Running standard deviation |
| runsum | Running sum |
| runvar1 | Running variance (n-1) |
| runvar | Running variance |
| samplegrid | Resample grid |
| sealevelpressure | Sea level pressure |
| seasavg | Seasonal average |
| seasmax | Seasonal maximum |
| seasmean | Seasonal mean |
| seasmin | Seasonal minimum |
| seaspctl | Seasonal percentiles |
| seasrange | Seasonal range |
| seasstd1 | Seasonal standard deviation (n-1) |
| seasstd | Seasonal standard deviation |
| seassum | Seasonal sum |
| seasvar1 | Seasonal variance (n-1) |
| seasvar | Seasonal variance |
| selcircle | Select cells inside a circle |
| selcode | Select parameters by code number |
| seldate | Select dates |
| selday | Select days |
| select | Select fields |
| selgridcell | Select grid cells |
| selgrid | Select grids |
| selhour | Select hours |
| selindexbox | Select an index box |
| sellevel | Select levels |
| sellevidx | Select levels by index |
| sellonlatbox | Select a longitude/latitude box |
| selltype | Select GRIB level types |
| selmonth | Select months |
| selmulti | Select multiple fields |
| selname | Select parameters by name |
| selparam | Select parameters by identifier |
| selregion | Select cells inside regions |
| selseason | Select seasons |
| selsmon | Select single month |
| selstdname | Select parameters by standard name |
| seltabnum | Select parameter table numbers |
| seltimeidx | Select timestep by index |
| seltimestep | Select timesteps |
| seltime | Select times |
| selyearidx | Select year by index |
| selyear | Select years |
| selzaxisname | Select z-axes by name |
| selzaxis | Select z-axes |
| seq | Create a time series |
| setattribute | Set attributes |
| setcalendar | Set calendar |
| setchunkspec | Specify chunking |
| setcindexbox | Set an index box to constant |
| setclonlatbox | Set a longitude/latitude box to constant |
| setcodetab | Set parameter code table |
| setcode | Set code number |
| setctomiss | Set constant to missing value |
| setdate | Set date |
| setday | Set day |
| setfilter | Specify filter |
| setgridarea | Set grid cell area |
| setgridcell | Set the value of a grid cell |
| setgridmask | Set grid mask |
| setgridtype | Set grid type |
| setgrid | Set grid |
| sethalo | Set the bounds of a field |
| setlevel | Set level |
| setltype | Set GRIB level type |
| setmaxsteps | Set max timesteps |
| setmisstoc | Set missing value to constant |
| setmisstodis | Set missing value to distance-weighted average |
| setmisstonn | Set missing value to nearest neighbor |
| setmissval | Set a new missing value |
| setmiss | Set missing values |
| setmon | Set month |
| setname | Set variable name |
| setparam | Set parameter identifier |
| setpartabn | Set parameter table |
| setpartabp | Set parameter table |
| setprojparams | Set proj params |
| setreftime | Set reference time |
| setrtoc2 | Set range to constant others to constant2 |
| setrtoc | Set range to constant |
| setrtomiss | Set range to missing value |
| setstdname | Set standard name |
| settaxis | Set time axis |
| settbounds | Set time bounds |
| settime | Set time of the day |
| settunits | Set time units |
| setunit | Set variable unit |
| setvals | Set list of old values to new values |
| setvrange | Set valid range |
| setyear | Set year |
| setzaxis | Set z-axis |
| shaded | Shaded contour plot |
| shifttime | Shift timesteps |
| shiftx | Shift x |
| shifty | Shift y |
| showattribute | Show a global attribute or a variable attribute |
| showchunkspec | Show chunk specification |
| showcode | Show code numbers |
| showdate | Show date information |
| showfilter | Show filter specification |
| showformat | Show file format |
| showlevel | Show levels |
| showltype | Show GRIB level types |
| showmon | Show months |
| showname | Show variable names |
| showstdname | Show standard names |
| showtimestamp | Show timestamp |
| showtime | Show time information |
| showyear | Show years |
| sinfon | Short information listed by parameter name |
| sinfo | Short information listed by parameter identifier |
| sin | Sine |
| smooth9 | 9 point smoothing |
| smooth | Smooth grid points |
| sp2gp | Spectral to gridpoint |
| sp2sp | Spectral to spectral |
| splitcode | Split code numbers |
| splitdate | Splits a file into dates |
| splitday | Split days |
| splitgrid | Split grids |
| splithour | Split hours |
| splitlevel | Split levels |
| splitmon | Split months |
| splitname | Split variable names |
| splitparam | Split parameter identifiers |
| splitseas | Split seasons |
| splitsel | Split time selection |
| splittabnum | Split parameter table numbers |
| splityearmon | Split in years and months |
| splityear | Split years |
| splitzaxis | Split z-axes |
| sqrt | Square root |
| sqr | Square |
| stdatm | Create values for pressure and temperature for hydrostatic atmosphere |
| strbre | Strong breeze days index per time period |
| strgal | Strong gale days index per time period |
| strwin | Strong wind days index per time period |
| subc | Subtract a constant |
| subtrend | Subtract trend |
| sub | Subtract two fields |
| tan | Tangent |
| tee | Duplicate a data stream |
| timavg | Time average |
| timcor | Correlation over time |
| timcovar | Covariance over time |
| timcumsum | Cumulative sum over all timesteps |
| timfillmiss | Temporal filling of missing values |
| timmaxidx | Index of time maximum |
| timmax | Time maximum |
| timmean | Time mean |
| timminidx | Index of time minimum |
| timmin | Time minimum |
| timpctl | Time percentiles |
| timrange | Time range |
| timselavg | Time selection average |
| timselmax | Time selection maximum |
| timselmean | Time selection mean |
| timselmin | Time selection minimum |
| timselpctl | Time range percentiles |
| timselrange | Time selection range |
| timselstd1 | Time selection standard deviation (n-1) |
| timselstd | Time selection standard deviation |
| timselsum | Time selection sum |
| timselvar1 | Time selection variance (n-1) |
| timselvar | Time selection variance |
| timsort | Sort over the time |
| timstd1 | Time standard deviation (n-1) |
| timstd | Time standard deviation |
| timsum | Time sum |
| timvar1 | Time variance (n-1) |
| timvar | Time variance |
| topo | Create a field with topography |
| topvalue | Extract top level |
| trend | Trend |
| unpack | Unpack data |
| uv2dv_cfd | U and V wind to divergence |
| uv2dv | U and V wind to divergence and vorticity |
| uv2vr_cfd | U and V wind to relative vorticity |
| uvDestag | Destaggering of u/v wind components |
| varsavg | Variables average |
| varsmax | Variables maximum |
| varsmean | Variables mean |
| varsmin | Variables minimum |
| varsrange | Variables range |
| varsstd1 | Variables standard deviation (n-1) |
| varsstd | Variables standard deviation |
| varssum | Variables sum |
| varsvar1 | Variables variance (n-1) |
| varsvar | Variables variance |
| vct | Vertical coordinate table |
| vector | Vector arrows plot |
| verifygrid | Verify grid coordinates |
| vertavg | Vertical average |
| vertfillmiss | Vertical filling of missing values |
| vertmax | Vertical maximum |
| vertmean | Vertical mean |
| vertmin | Vertical minimum |
| vertrange | Vertical range |
| vertstd1 | Vertical standard deviation (n-1) |
| vertstd | Vertical standard deviation |
| vertsum | Vertical sum |
| vertvar1 | Vertical variance (n-1) |
| vertvar | Vertical variance |
| wct | Windchill temperature |
| xsinfop | Extra short information listed by parameter identifier |
| xsinfo | Extra short information listed by parameter name |
| ydayadd | Add multi-year daily time series |
| ydayavg | Multi-year daily average |
| ydaydiv | Divide multi-year daily time series |
| ydaymax | Multi-year daily maximum |
| ydaymean | Multi-year daily mean |
| ydaymin | Multi-year daily minimum |
| ydaymul | Multiply multi-year daily time series |
| ydaypctl | Multi-year daily percentiles |
| ydayrange | Multi-year daily range |
| ydaystd1 | Multi-year daily standard deviation (n-1) |
| ydaystd | Multi-year daily standard deviation |
| ydaysub | Subtract multi-year daily time series |
| ydaysum | Multi-year daily sum |
| ydayvar1 | Multi-year daily variance (n-1) |
| ydayvar | Multi-year daily variance |
| ydrunavg | Multi-year daily running average |
| ydrunmax | Multi-year daily running maximum |
| ydrunmean | Multi-year daily running mean |
| ydrunmin | Multi-year daily running minimum |
| ydrunpctl | Multi-year daily running percentiles |
| ydrunstd1 | Multi-year daily running standard deviation (n-1) |
| ydrunstd | Multi-year daily running standard deviation |
| ydrunsum | Multi-year daily running sum |
| ydrunvar1 | Multi-year daily running variance (n-1) |
| ydrunvar | Multi-year daily running variance |
| yearadd | Add yearly time series |
| yearavg | Yearly average |
| yeardiv | Divide yearly time series |
| yearmaxidx | Index of yearly maximum |
| yearmax | Yearly maximum |
| yearmean | Yearly mean |
| yearminidx | Index of yearly minimum |
| yearmin | Yearly minimum |
| yearmonmean | Yearly mean from monthly data |
| yearmul | Multiply yearly time series |
| yearpctl | Yearly percentiles |
| yearrange | Yearly range |
| yearstd1 | Yearly standard deviation (n-1) |
| yearstd | Yearly standard deviation |
| yearsub | Subtract yearly time series |
| yearsum | Yearly sum |
| yearvar1 | Yearly variance (n-1) |
| yearvar | Yearly variance |
| yhouradd | Add multi-year hourly time series |
| yhouravg | Multi-year hourly average |
| yhourdiv | Divide multi-year hourly time series |
| yhourmax | Multi-year hourly maximum |
| yhourmean | Multi-year hourly mean |
| yhourmin | Multi-year hourly minimum |
| yhourmul | Multiply multi-year hourly time series |
| yhourrange | Multi-year hourly range |
| yhourstd1 | Multi-year hourly standard deviation (n-1) |
| yhourstd | Multi-year hourly standard deviation |
| yhoursub | Subtract multi-year hourly time series |
| yhoursum | Multi-year hourly sum |
| yhourvar1 | Multi-year hourly variance (n-1) |
| yhourvar | Multi-year hourly variance |
| ymonadd | Add multi-year monthly time series |
| ymonavg | Multi-year monthly average |
| ymondiv | Divide multi-year monthly time series |
| ymoneq | Compare time series with Equal |
| ymonge | Compares if time series with GreaterEqual |
| ymongt | Compares if time series with GreaterThan |
| ymonle | Compare time series with LessEqual |
| ymonlt | Compares if time series with LessThan |
| ymonmax | Multi-year monthly maximum |
| ymonmean | Multi-year monthly mean |
| ymonmin | Multi-year monthly minimum |
| ymonmul | Multiply multi-year monthly time series |
| ymonne | Compare time series with NotEqual |
| ymonpctl | Multi-year monthly percentiles |
| ymonrange | Multi-year monthly range |
| ymonstd1 | Multi-year monthly standard deviation (n-1) |
| ymonstd | Multi-year monthly standard deviation |
| ymonsub | Subtract multi-year monthly time series |
| ymonsum | Multi-year monthly sum |
| ymonvar1 | Multi-year monthly variance (n-1) |
| ymonvar | Multi-year monthly variance |
| yseasadd | Add multi-year seasonal time series |
| yseasavg | Multi-year seasonal average |
| yseasdiv | Divide multi-year seasonal time series |
| yseaseq | Compare time series with Equal |
| yseasge | Compares if time series with GreaterEqual |
| yseasgt | Compares if time series with GreaterThan |
| yseasle | Compare time series with LessEqual |
| yseaslt | Compares if time series with LessThan |
| yseasmax | Multi-year seasonal maximum |
| yseasmean | Multi-year seasonal mean |
| yseasmin | Multi-year seasonal minimum |
| yseasmul | Multiply multi-year seasonal time series |
| yseasne | Compare time series with NotEqual |
| yseaspctl | Multi-year seasonal percentiles |
| yseasrange | Multi-year seasonal range |
| yseasstd1 | Multi-year seasonal standard deviation (n-1) |
| yseasstd | Multi-year seasonal standard deviation |
| yseassub | Subtract multi-year seasonal time series |
| yseassum | Multi-year seasonal sum |
| yseasvar1 | Multi-year seasonal variance (n-1) |
| yseasvar | Multi-year seasonal variance |
| zaxisdes | Z-axis description |
| zonavg | Zonal average |
| zonkurt | Zonal kurtosis |
| zonmax | Zonal maximum |
| zonmean | Zonal mean |
| zonmedian | Zonal median |
| zonmin | Zonal minimum |
| zonpctl | Zonal percentiles |
| zonrange | Zonal range |
| zonskew | Zonal skewness |
| zonstd1 | Zonal standard deviation (n-1) |
| zonstd | Zonal standard deviation |
| zonsum | Zonal sum |
| zonvar1 | Zonal variance (n-1) |
| zonvar | Zonal variance |