projection_query
Added by kunal bali almost 7 years ago
Dear CDO users,
I have been using these code to convert curvlinear to rectilinear. And these code have been working perfectly fine but not for the attached file. So could you please let me know how to convert/or to assign coordinates of the given file into rectilinear file.
load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/gsn_code.ncl"
load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/gsn_csm.ncl"
load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/contributed.ncl"
load "$NCARG_ROOT/lib/ncarg/nclscripts/esmf/ESMF_regridding.ncl"
;
OutFileName = "cur_to_rect_day_01.nc"
;
f = addfile("all.nc", "r")
lat2d = f->latitude
lon2d = f->longitude
;
Opt = True
Opt@InterpMethod = "bilinear" ; default
Opt@ForceOverwrite = True
Opt@PrintTimings = True
;
rectilinear_to_SCRIP(OutFileName,lat2d,lon2d,Opt)
<pre>
<pre>
cdo -f nc -setgrid,scrip_grid.nc out.nc new_out.nc
<pre>
Thank You
Kunal Bali
Replies (8)
RE: projection_query - Added by Karin Meier-Fleischer almost 7 years ago
Hi Kunak,
I think, this script haven't had run ever. I wouldn't recommend to mix NCL and CDO in that way.
First, you have to add the coordinates attribute to your variables
ncatted -O -a coordinates,Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04,c,c,"longitude latitude" \
-a coordinates,Aerosol_Cloud_Fraction_Land_mod04,c,c,"longitude latitude" \
all.nc all_coordinates.ncThen you can compute the weights
cdo genbil,r180x90 all_coordinates.nc weights.nc
Do the remapping
cdo remap,r180x90,weights.nc all_coordinates.nc out_kunal.nc
Create the plot of first timestep for e.g. Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04
cdo grfill,device="png" -seltimestep,1 -selname,Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04 out_kunal.nc plot_out_kunal
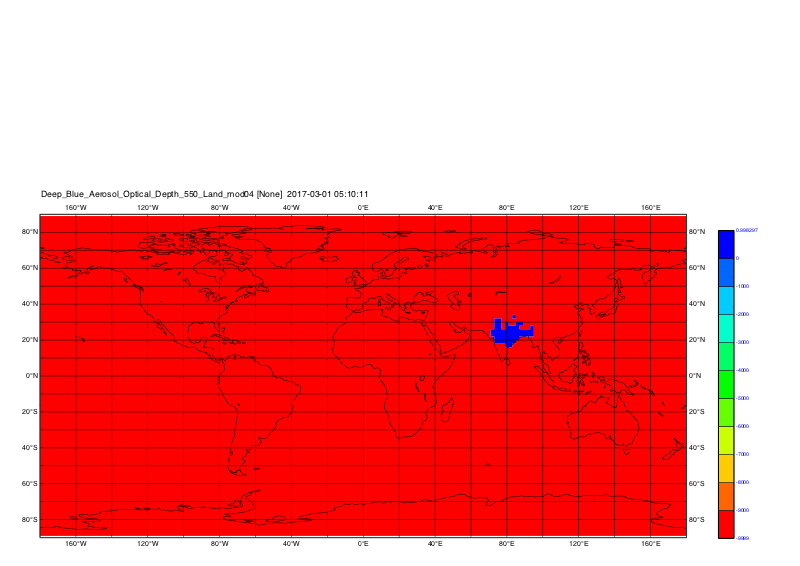
-Karin
RE: projection_query - Added by kunal bali almost 7 years ago
Sir,
Thanks for the input. I have 2 query regarding this issue:
1.) The final output (out_kunal.nc) is not matching with the input file (all.nc) WITH TIME STEPS. I mean projection is different.
2.) The all.nc file was created from the intial attached file (MOD04.2017030105-2017032304.nc). That was created by the cdo splitday and then the cdo mergetime code (just wanted to check whether cdo working on this projection or not).
But when I try your code on the attached file MOD04.2017030105-2017032304.nc then it's not reading the Longitude_mod04 Latitude_mod04.
RE: projection_query - Added by Karin Meier-Fleischer almost 7 years ago
Please, take a look at your data!
1) They have exactly the same time steps:
cdo showtimestamp all.nc2017-03-01T05:10:11 2017-03-10T05:05:11 2017-03-15T05:20:11 2017-03-19T05:00:11 2017-03-23T04:35:11
cdo showtimestamp: Processed 4 variables over 5 timesteps ( 0.00s 20MB )
cdo showtimestamp out_kunal.nc2017-03-01T05:10:11 2017-03-10T05:05:11 2017-03-15T05:20:11 2017-03-19T05:00:11 2017-03-23T04:35:11
cdo showtimestamp: Processed 2 variables over 5 timesteps ( 0.00s 20MB )
2) And again, take a look at your data.
The variables in this file MOD04.2017030105-2017032304.nc have already a coordinates attribute
Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04:coordinates = "Latitude_mod04, Longitude_mod04" ;
Aerosol_Cloud_Fraction_Land_mod04:coordinates = "Latitude_mod04, Longitude_mod04" ;
but the variables Latitude_mod04 and Longitude_mod04 are missing in the file.
-Karin
RE: projection_query - Added by kunal bali almost 7 years ago
Yes, you are right sir, both the files have same time steps. But if you look at the 4rth and 5th timestep then both the files have different projections. Both the files have data at different lat Lon. e.g I have attached the figures of 4rth time step for the files all.nc and out_kunal. <pre>
| all.png (312 KB) all.png | |||
| out_kunal.png (298 KB) out_kunal.png |
RE: projection_query - Added by Karin Meier-Fleischer almost 7 years ago
Yupp, the latitude and longitude coordinate variables are time depending. The time dependency must be
deleted which means you have to create one file per time step and delete the time dimension in
latitude and longitude. In my opinion CDO can't do that, so there comes NCL back into the game to
do that.
Here is a short Korn Shell script which generates the NCL script to read the data all_coordinates.nc
which we have created earlier, select the time step, delete the time in latitude and longitude and
calls CDO to do the remapping (and some plots, too ;). You can do the remapping in NCL without
CDO's, too, but we're here in the CDO forum, so let's do it this way.
myscript.ksh
#!/bin/ksh
cat << EOF > script.ncl
f = addfile("all_coordinates.nc","r")
time = f->time
ntime = dimsizes(f->time)
lat = f->latitude
lon = f->longitude
do t=0,ntime-1
system("rm -rf out_all_coordinates_delete_time_in_lonlat"+t+".nc")
outf := addfile("out_all_coordinates_delete_time_in_lonlat"+t+".nc","c")
outf->latitude = lat(t,:,:)
outf->longitude = lon(t,:,:)
outf->Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04 = f->Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04(t:t,:,:)
outf->Aerosol_Cloud_Fraction_Land_mod04 = f->Aerosol_Cloud_Fraction_Land_mod04(t:t,:,:)
delete(outf)
system("cdo -O genbil,r180x90 out_all_coordinates_delete_time_in_lonlat"+t+".nc weights_"+t+".nc")
system("cdo -O remap,r180x90,weights_"+t+".nc out_all_coordinates_delete_time_in_lonlat"+t+".nc out_kunal_"+t+".nc")
system("cdo grfill,device="+str_get_dq()+"png"+str_get_dq()+",min=0,max=1.0,interval=0.05,lon_min=60,lon_max=100,lat_min=10,lat_max=40 -selname,Aerosol_Cloud_Fraction_Land_mod04 out_kunal_"+t+".nc plot_kunal_all_coord_"+t)
end do
EOF
ncl script.ncl
exit
Run the script
ksh myscript.ksh
The plots are
Time step 1: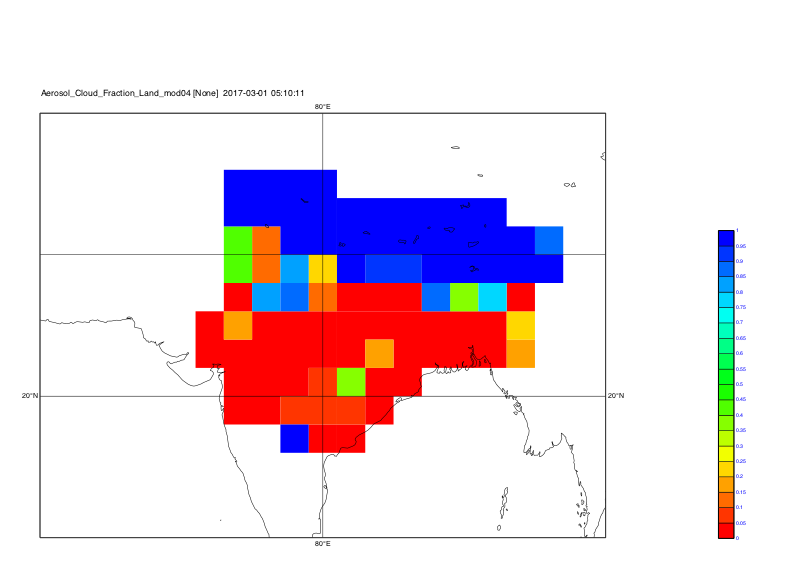
Time step 2: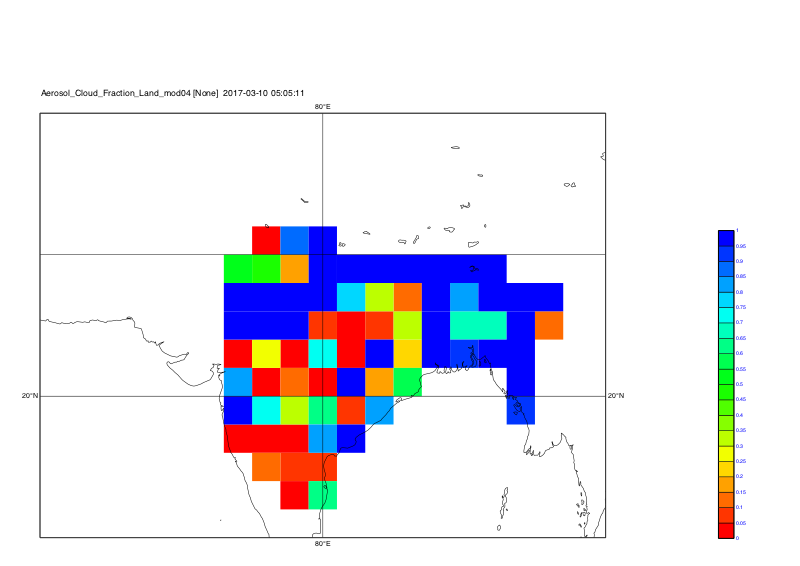
Time step 3: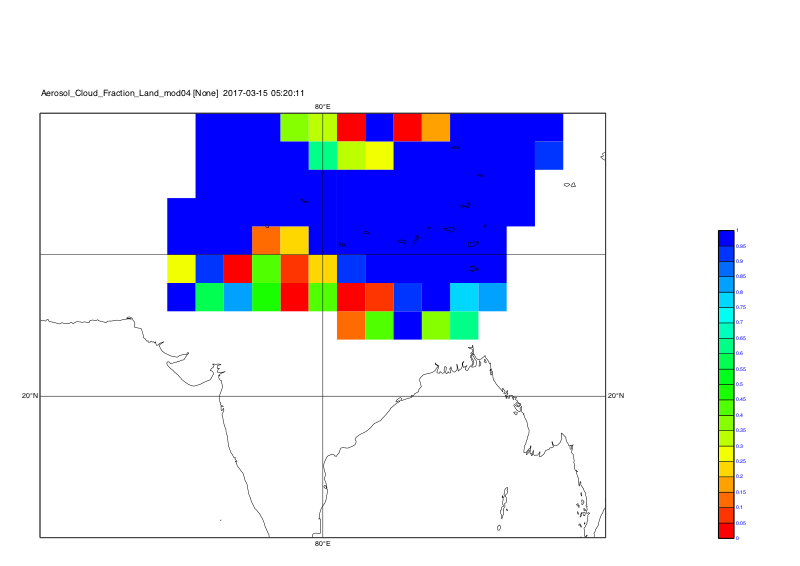
Time step 4: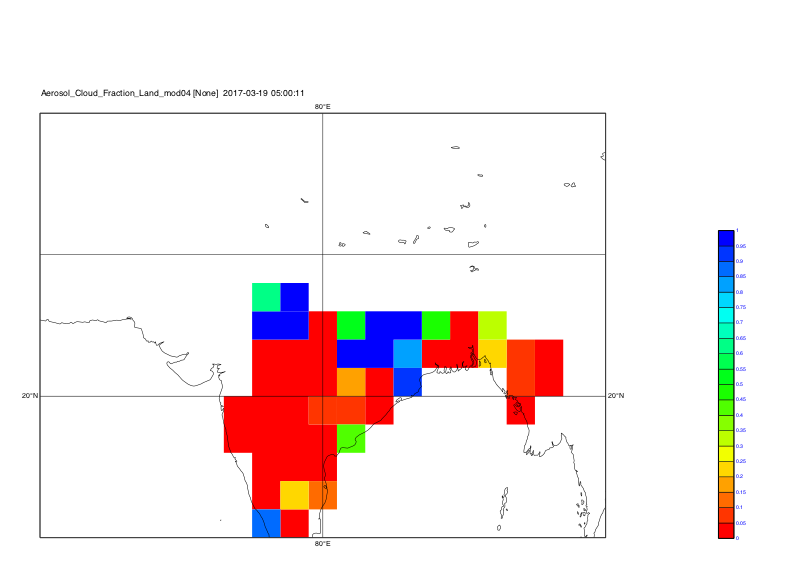
Time step 5: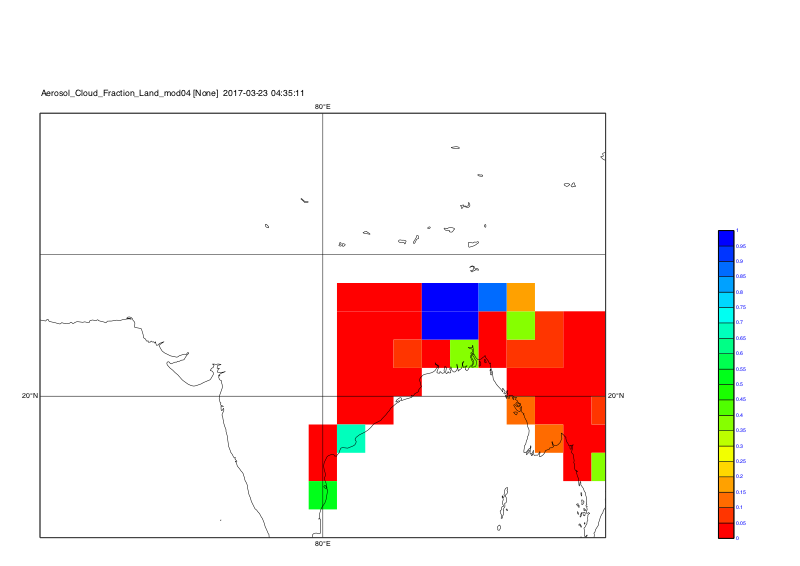
-Karin
RE: projection_query - Added by kunal bali almost 7 years ago
Thank You, sir. It worked perfectly.
I have one question regarding Latitude_mod04, Longitude_mod04 which I asked you earlier and you mentioned that the variables Latitude_mod04 and Longitude_mod04 are missing in the file. yes, you are right.
But when I split the data using
cdo splitday inputfile.nc day_ and then merge the time again using <pre> cod mergetime day*.nc all.nc and then I run the script myscript.ksh. This time I don't get any error regarding Latitude_mod04 and Longitude_mod04. So, is that mean CDO replaced/or deleted the Latitude_mod04 and Longitude_mod04 automatically? how did CDO work in this case? However, This process is working perfectly. I am getting all the projection correctly.
RE: projection_query - Added by Karin Meier-Fleischer almost 7 years ago
When you do 'cdo splitday' CDO will return a warning that Latitude_mod04 and Longitude_mod04 is missing
Warning (cdf_scan_var_attr) : NetCDF: Variable not found - >Latitude_mod04< Warning (cdf_scan_var_attr) : NetCDF: Variable not found - >Longitude_mod04<
and it will remove the coordinates attribute from the variables. Have a look at your data all.nc with ncdump.
Here, I called it myall.nc because your original file is already named all.nc.
ncdump -h myall.nc
netcdf myall {
dimensions:
time = UNLIMITED ; // (5 currently)
Cell_Across_Swath_mod04 = 135 ;
Cell_Along_Swath_mod04 = 204 ;
variables:
double time(time) ;
time:standard_name = "time" ;
time:long_name = "initial time of scan" ;
time:units = "Seconds since 1993-1-1 00:00:00.0 0" ;
time:calendar = "standard" ;
time:axis = "T" ;
float latitude(time, Cell_Along_Swath_mod04, Cell_Across_Swath_mod04) ;
latitude:long_name = "latitude" ;
latitude:units = "degrees_north" ;
latitude:_FillValue = -999.f ;
latitude:missing_value = -999.f ;
latitude:hdfeos_name = "Latitude" ;
float longitude(time, Cell_Along_Swath_mod04, Cell_Across_Swath_mod04) ;
longitude:long_name = "longitude" ;
longitude:units = "degrees_east" ;
longitude:_FillValue = -999.f ;
longitude:missing_value = -999.f ;
longitude:hdfeos_name = "Longitude" ;
float Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04(time, Cell_Along_Swath_mod04, Cell_Across_Swath_mod04) ;
Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04:long_name = "AOT at 0.55 micron for land with all quality data (Quality flag=1,2,3)" ;
Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04:units = "None" ;
Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04:_FillValue = -9999.f ;
Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04:missing_value = -9999.f ;
Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04:hdfeos_name = "Deep_Blue_Aerosol_Optical_Depth_550_Land" ;
Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04:Geolocation_Pointer = "Internal geolocation arrays" ;
Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04:Cell_Across_Swath_Sampling = 1, 1354, 10 ;
Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04:Cell_Along_Swath_Sampling = 1, 2031, 10 ;
Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04:Parameter_Type = "Output" ;
Deep_Blue_Aerosol_Optical_Depth_550_Land_mod04:_FillValue_original = -9999s ;
float Aerosol_Cloud_Fraction_Land_mod04(time, Cell_Along_Swath_mod04, Cell_Across_Swath_mod04) ;
Aerosol_Cloud_Fraction_Land_mod04:long_name = "Cloud fraction from Land aerosol cloud mask from retrieved and overcast pixels not including cirrus mask" ;
Aerosol_Cloud_Fraction_Land_mod04:units = "None" ;
Aerosol_Cloud_Fraction_Land_mod04:_FillValue = -9999.f ;
Aerosol_Cloud_Fraction_Land_mod04:missing_value = -9999.f ;
Aerosol_Cloud_Fraction_Land_mod04:hdfeos_name = "Aerosol_Cloud_Fraction_Land" ;
Aerosol_Cloud_Fraction_Land_mod04:Geolocation_Pointer = "Internal geolocation arrays" ;
Aerosol_Cloud_Fraction_Land_mod04:Cell_Across_Swath_Sampling = 1, 1354, 10 ;
Aerosol_Cloud_Fraction_Land_mod04:Cell_Along_Swath_Sampling = 1, 2031, 10 ;
Aerosol_Cloud_Fraction_Land_mod04:Parameter_Type = "Output" ;
Aerosol_Cloud_Fraction_Land_mod04:_FillValue_original = -9999s ;
// global attributes:
:CDI = "Climate Data Interface version 1.9.2 (http://mpimet.mpg.de/cdi)" ;
:Conventions = "None" ;
:history = "Tue Aug 14 10:01:43 2018: cdo mergetime day_01.nc day_10.nc day_15.nc day_19.nc day_23.nc myall.nc\n",
"Tue Aug 14 10:01:43 2018: cdo splitday MOD04.2017030105-2017032304.nc day_" ;
:creation_date = "Mon Aug 13 13:57:50 IST 2018" ;
:NCL_version = "Any version of NCL >= 5.2.0 (4/2010)" ;
:source_file = "MOD04_L2AOT HDF-EOS files" ;
:title = "MOD04_L2AOT HDF-EOS: time dimension added to selected variables" ;
:CDO = "Climate Data Operators version 1.9.2 (http://mpimet.mpg.de/cdo)" ;
}
-Karin