CDO python bindings do not work in jupyterhub on MISTRAL
Added by Diego Jimenez over 6 years ago
I am trying to process a netCDF file in a Jupyter notebook on MISTRAL, but it fails always, no matter what operator I use I get an unknown error that the file cannot be accessed. I have tested with the command cdo.infon for different files (in my home directory, in the same directory where the notebook is or in my work directory) to no avail. Is this a limitation of the jupyter server in MISTRAL? Or there are problems with permissions to read files
The imports are
import numpy as np # Numpy from cdo import * cdo=Cdo()
And the failing code and error message
hcon=cdo.timmean(input='heat_content_anomaly_0-700_yearly.nc',options='-f nc4') Error in calling operator timmean with: >>> cdo -O -f nc4 timmean heat_content_anomaly_0-700_yearly.nc /tmp/cdoPywr3P2L<<< STDOUT: STDERR:cdo timmean: Open failed on >heat_content_anomaly_0-700_yearly.nc< Unknown Error
Replies (14)
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Ralf Mueller over 6 years ago
hi Diego!
simple: the file is not where cdo expects it. try an absolute path instead to avoid such errors, esp using jupyter
hth
ralf
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Diego Jimenez over 6 years ago
I tried with absolute paths and error is still there, same imports,
direc='/work/mh0469/m300556/CMIP5catdata/MPI-M/MPI-ESM1.1/p0/' frsln='rsln_mon_piControl_r001_i1_p0_MPI-ESM1.1_1850-3850.nc' rsln=cdo.infon(input=direc+frsln)
Error in calling operator infon with: >>> cdo -O infon /work/mh0469/m300556/CMIP5catdata/MPI-M/MPI-ESM1.1/p1/rsut_mon_piControl_r001_i1_p1_MPI-ESM1.1_1850-3850.nc<<< STDOUT: STDERR:cdo infon: Open failed on >/work/mh0469/m300556/CMIP5catdata/MPI-M/MPI-ESM1.1/p1/rsut_mon_piControl_r001_i1_p1_MPI-ESM1.1_1850-3850.nc< Unknown Error
When I use the netcdf4 module to load a file with the Dataset class, there is no problem in loading with absolute or relative paths.
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Diego Jimenez over 6 years ago
I suppose that I should contact dkrz.
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Ralf Mueller over 6 years ago
I can run this in an interactive python shell
import cdo c = cdo.Cdo() print(c.infov(input=' -seltimestep,1 /work/mh0469/m300556/CMIP5catdata/MPI-M/MPI-ESM1.1/p1/rsut_mon_piControl_r001_i1_p1_MPI-ESM1.1_1850-3850.nc')) ['-1 : Date Time Level Gridsize Miss : Minimum Mean Maximum : Parameter name', '1 : 1850-01-01 00:00:00 0 18432 0 : -355.16 -119.01 -0.0019531 : rsut']
I will switch to the jupyter service ....
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Karin Meier-Fleischer over 6 years ago
Hi Diego,
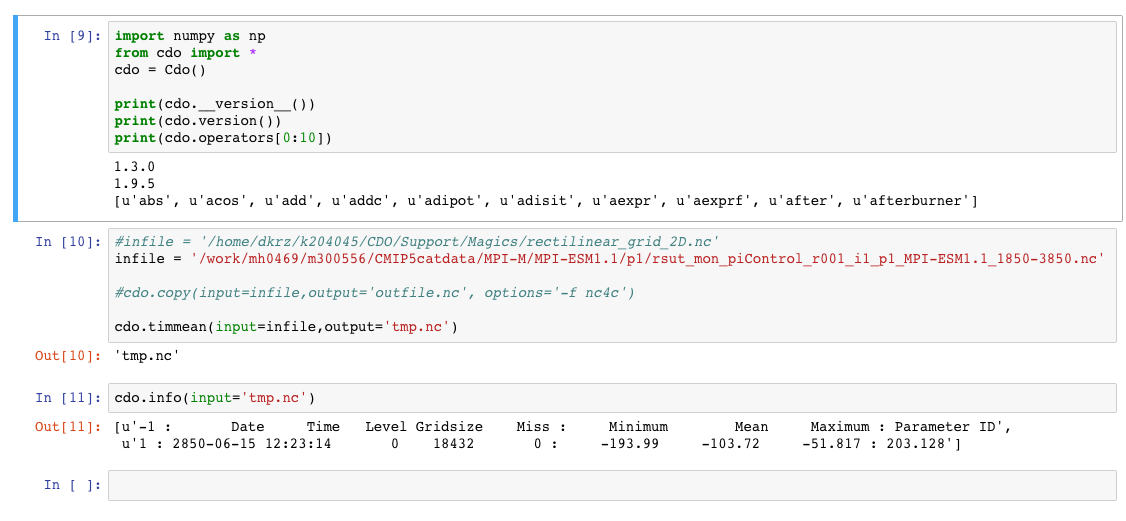
at mistral the following is working fine within the notebook:
import numpy as np from cdo import * cdo = Cdo() print(cdo.version()) infile = '/work/mh0469/m300556/CMIP5catdata/MPI-M/MPI-ESM1.1/p1/rsut_mon_piControl_r001_i1_p1_MPI-ESM1.1_1850-3850.nc' cdo.timmean(input=infile,output='tmp.nc') cdo.info(input='tmp.nc')
Output: 1.9.5 [u'-1 : Date Time Level Gridsize Miss : Minimum Mean Maximum : Parameter ID', u'1 : 2850-06-15 12:23:14 0 18432 0 : -193.99 -103.72 -51.817 : 203.128']-Karin
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Ralf Mueller over 6 years ago
+1 @Karin
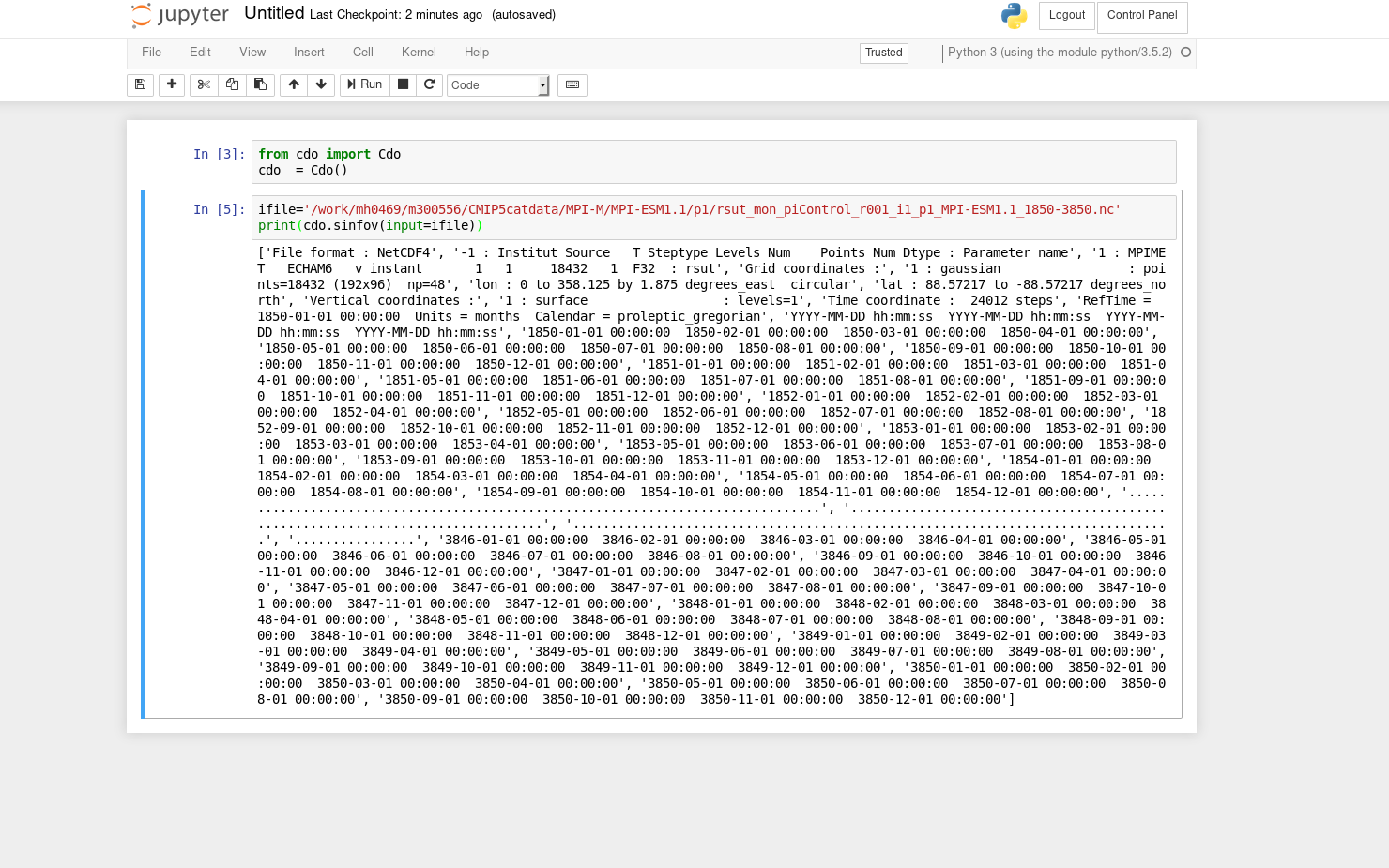
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Diego Jimenez over 6 years ago
Thank you Ralf and Karin,
Yes, in mistral with Ipython everything works well. But running the same code reserving resources through jupyterhub.dkrz.de is what is failing. Then, that means that probably is a problem of jupyterhub and not of cdo.py which is a good news 
Cheers,
Diego.
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Ralf Mueller over 6 years ago
are you in the office? I could stop by to have a look at your notebook...
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Diego Jimenez over 6 years ago
I think that the problem is I am using python 2.7.12. Yes I am at the office. B429
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Karin Meier-Fleischer over 6 years ago
I used https://jupyterhub.dkrz.de/ to run the notebook input at mistral.
You can find the notebook at /home/dkrz/k204045/CDO/Support/Python/cdo_notebook_test.ipynb
-Karin
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Karin Meier-Fleischer over 6 years ago
I'm using python/2.7.12, too.
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Diego Jimenez over 6 years ago
Here are the results in two separate notebooks. I am not doing but what you have in your notebook
RE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Karin Meier-Fleischer over 6 years ago
Please, don't mix python and cdo syntax.
import numpy as np
from cdo import *
import matplotlib.pyplot as plt
from netCDF4 import Dataset as open_ncfile
cdo = Cdo()
direc = '/work/mh0469/m300556/CMIP5catdata/MPI-M/MPI-ESM1.1/p0/'
frsln = 'rsln_mon_piControl_r001_i1_p0_MPI-ESM1.1_1850-3850.nc'
ftsur = 'ts_mon_piControl_r001_i1_p0_MPI-ESM1.1_1850-3850.nc'
cdo.timmean(input='-select,name=N '+direc+frsln,output='tmp1.nc',returnCDF=True)
cdo.timstd(input='-select,name=N '+direc+frsln,output='tmp2.nc',returnCDF=True)
nc1 = open_ncfile('tmp1.nc')
mN = nc1.variables['N'][0,:,:]
nc2 = open_ncfile('tmp2.nc')
sN = nc2.variables['N'][0,:,:]
print(mN)
print('---------------------')
print(sN)
-KarinRE: CDO python bindings do not work in jupyterhub on MISTRAL - Added by Karin Meier-Fleischer over 6 years ago
Ok, your notebook above should be:
import numpy as np
from cdo import *
import matplotlib.pyplot as plt
from netCDF4 import Dataset as open_ncfile
cdo = Cdo()
direc = '/work/mh0469/m300556/CMIP5catdata/MPI-M/MPI-ESM1.1/p0/'
frsln = 'rsln_mon_piControl_r001_i1_p0_MPI-ESM1.1_1850-3850.nc'
ftsur = 'ts_mon_piControl_r001_i1_p0_MPI-ESM1.1_1850-3850.nc'
cdo.timmean(input='-select,name=N '+direc+frsln,output='tmp1.nc',returnCDF=True)
cdo.timstd(input='-select,name=N '+direc+frsln,output='tmp2.nc',returnCDF=True)
nc1 = open_ncfile('tmp1.nc')
nc2 = open_ncfile('tmp2.nc')
mN = nc1.variables['N'][0,:,:]
sN = nc2.variables['N'][0,:,:]
cdo.runmean(12,input='-div -sub '+direc+frsln+' -timmean '+direc+frsln+' -timstd '+direc+frsln,\
output='tmp3.nc',options='-f nc4',returnCDF=True)
cdo.runmean(12,input='-div -sub '+direc+ftsur+' -timmean '+direc+ftsur+' -timstd '+direc+ftsur,\
output='tmp4.nc',options='-f nc4',returnCDF=True)
nc3 = open_ncfile('tmp3.nc')
nc4 = open_ncfile('tmp4.nc')
N = nc3.variables['N'][0,:,:]
T = nc4.variables['T'][0,:,:]
print(N)
print('---------------------')
print(T)
[[ 0.01243716 0.01225562 0.01298991 ..., 0.00922786 0.01142903 0.01117445] [ 0.02401677 0.02658143 0.02832476 ..., 0.01943446 0.02033182 0.02166243] [ 0.03806251 0.0375117 0.03549416 ..., 0.03068122 0.03205523 0.03420843] ..., [-0.02417499 -0.02061274 -0.02112462 ..., -0.02755507 -0.03012871 -0.02446408] [-0.02941976 -0.02559732 -0.02466218 ..., -0.02980293 -0.02855076 -0.02911846] [ 0.00821737 0.00986399 0.011079 ..., 0.00390146 0.00417242 0.00682933]] --------------------- [[-0.03028085 -0.03144855 -0.03151836 ..., -0.03119568 -0.03031179 -0.02991755] [-0.03851259 -0.04040512 -0.04350967 ..., -0.03002559 -0.03401553 -0.03769588] [-0.04358529 -0.05069558 -0.04870506 ..., -0.03333439 -0.0375292 -0.03746813] ..., [ 0.08453191 0.07819933 0.07968152 ..., 0.10025381 0.10227605 0.08719432] [ 0.09559853 0.08886145 0.08885103 ..., 0.09355759 0.08899032 0.09203639] [ 0.01200529 0.00943254 0.00686293 ..., 0.02022018 0.01935472 0.01476929]]-Karin