segmentation fault
Added by Hanne Byre over 6 years ago
Hei!
I have selected an area by lat,lon, put a variable in sector 1 (sector=1-8(dim)) to zero. Now I want to put this area back again to the global file, but I get this error message "Error (cdf_get_vara_double) : NetCDF: HDF error
Segmentation fault"
I have typed "cdo -add -setclonlatbox,0,7.5,37.5,67.5,97.5 NOx-em-anthro_input4MIPs_emissions_CIP_CEDS-2017-05-18_gn_200001-201412.nc -setmisstoc,0 -enlargegrid,NOx-em-anthro_input4MIPs_emissions_CIP_CEDS-2017-05-18_gn_200001-201412.nc ne2windia.nc nyfil.nc"
where "NOx-em-anthro_input4MIPs_emissions_CIP_CEDS-2017-05-18_gn_200001-201412.nc" is my global file,
and "ne2windia.nc" is the file that contains only the selected area, where sector 1, var1 it put to zero, rest is kept the same as the global file.
nyfil.nc is the new file where the subset is added.
ncdump -h NOx-em-anthro_input4MIPs_emissions_CIP_CEDS-2017-05-18_gn_200001-201412.nc
dimensions:
lon = 720 ;
lat = 360 ;
sector = 8 ;
time = UNLIMITED ; // (180 currently)
bound = 2 ;
variables:
double lon(lon) ;
lon:units = "degrees_east" ;
lon:long_name = "longitude" ;
lon:axis = "X" ;
lon:bounds = "lon_bnds" ;
lon:modulo = 360. ;
lon:realtopology = "circular" ;
lon:standard_name = "longitude" ;
lon:topology = "circular" ;
double lat(lat) ;
lat:units = "degrees_north" ;
lat:long_name = "latitude" ;
lat:axis = "Y" ;
lat:bounds = "lat_bnds" ;
lat:realtopology = "linear" ;
lat:standard_name = "latitude" ;
int sector(sector) ;
sector:long_name = "sector" ;
sector:bounds = "sector_bnds" ;
sector:ids = "0: Agriculture; 1: Energy; 2: Industrial; 3: Transportation; 4: Residential, Commercial, Other; 5: Solvents production and application; 6: Waste; 7: International Shipping" ;
double time(time) ;
time:units = "days since 1750-01-01 0:0:0" ;
time:long_name = "time" ;
time:calendar = "365_day" ;
time:axis = "T" ;
time:bounds = "time_bnds" ;
time:realtopology = "linear" ;
time:standard_name = "time" ;
float NOx_em_anthro(time, sector, lat, lon) ;
NOx_em_anthro:units = "kg m-2 s-1" ;
NOx_em_anthro:_FillValue = 1.e+20f ;
NOx_em_anthro:long_name = "NOx Anthropogenic Emissions" ;
NOx_em_anthro:cell_methods = "time: mean" ;
NOx_em_anthro:missing_value = 1.e+20f ;
double lat_bnds(lat, bound) ;
double lon_bnds(lon, bound) ;
double time_bnds(time, bound) ;
double sector_bnds(sector, bound) ;
ncdump -h ne2windia.nc
dimensions:
time = UNLIMITED ; // (180 currently)
sector = 8 ;
lat = 60 ;
lon = 60 ;
bound = 2 ;
variables:
float NOx_em_anthro(time, sector, lat, lon) ;
NOx_em_anthro:_FillValue = 1.e+20f ;
NOx_em_anthro:cell_methods = "time: mean" ;
NOx_em_anthro:long_name = "NOx Anthropogenic Emissions" ;
NOx_em_anthro:missing_value = 1.e+20f ;
NOx_em_anthro:units = "kg m-2 s-1" ;
double lon(lon) ;
lon:units = "degrees_east" ;
lon:long_name = "longitude" ;
lon:axis = "X" ;
lon:bounds = "lon_bnds" ;
lon:modulo = 360. ;
lon:realtopology = "circular" ;
lon:standard_name = "longitude" ;
lon:topology = "circular" ;
double lat(lat) ;
lat:units = "degrees_north" ;
lat:long_name = "latitude" ;
lat:axis = "Y" ;
lat:bounds = "lat_bnds" ;
lat:realtopology = "linear" ;
lat:standard_name = "latitude" ;
int sector(sector) ;
sector:long_name = "sector" ;
sector:bounds = "sector_bnds" ;
sector:ids = "0: Agriculture; 1: Energy; 2: Industrial; 3: Transportation; 4: Residential, Commercial, Other; 5: Solvents production and application; 6: Waste; 7: International Shipping" ;
double time(time) ;
time:units = "days since 1750-01-01 0:0:0" ;
time:long_name = "time" ;
time:calendar = "365_day" ;
time:axis = "T" ;
time:bounds = "time_bnds" ;
time:realtopology = "linear" ;
time:standard_name = "time" ;
double lat_bnds(lat, bound) ;
double lon_bnds(lon, bound) ;
double time_bnds(time, bound) ;
double sector_bnds(sector, bound) ;
ncdump -h nyfil.nc
dimensions:
lon = 720 ;
nb2 = 2 ;
lat = 360 ;
sector = 8 ;
time = UNLIMITED ; // (1 currently)
variables:
double lon(lon) ;
lon:standard_name = "longitude" ;
lon:long_name = "longitude" ;
lon:units = "degrees_east" ;
lon:axis = "X" ;
lon:bounds = "lon_bnds" ;
double lon_bnds(lon, nb2) ;
double lat(lat) ;
lat:standard_name = "latitude" ;
lat:long_name = "latitude" ;
lat:units = "degrees_north" ;
lat:axis = "Y" ;
lat:bounds = "lat_bnds" ;
double lat_bnds(lat, nb2) ;
double sector(sector) ;
sector:long_name = "sector" ;
sector:axis = "Z" ;
sector:bounds = "sector_bnds" ;
double sector_bnds(sector, nb2) ;
double time(time) ;
time:standard_name = "time" ;
time:long_name = "time" ;
time:bounds = "time_bnds" ;
time:units = "days since 1750-01-01 00:00:00" ;
time:calendar = "365_day" ;
double time_bnds(time, nb2) ;
float NOx_em_anthro(time, sector, lat, lon) ;
NOx_em_anthro:long_name = "NOx Anthropogenic Emissions" ;
NOx_em_anthro:units = "kg m-2 s-1" ;
NOx_em_anthro:_FillValue = 1.e+20f ;
NOx_em_anthro:missing_value = 1.e+20f ;
NOx_em_anthro:cell_methods = "time: mean" ;
As you can see, there is something weird happening to the time in the newfile "nyfil.nc"
It this the problem? And how can i solve it ?
-Hanne Eline
Replies (15)
RE: segmentation fault - Added by Ralf Mueller over 6 years ago
hi Hanne!
for Further help, it would be good to have to dat available. Could upload at least some sample of it, please?
I am a little confused by your call: You add two streams:
-setclonlatbox,0,7.5,37.5,67.5,97.5 NOx-em-anthro_input4MIPs_emissions_CIP_CEDS-2017-05-18_gn_200001-201412.nc-setmisstoc,0 -enlargegrid,NOx-em-anthro_input4MIPs_emissions_CIP_CEDS-2017-05-18_gn_200001-201412.nc ne2windia.nc
Stream 1 is a non-global grid, but stream 2 is global - so the call should fail with an error IMO.
cheers
ralf
RE: segmentation fault - Added by Hanne Byre over 6 years ago
Thanks! I've uploaded the data. I think there is a problem with the time, since the variable NOx_em_anthro depends on (time, sector, lat, lon), and in have only specified which lat/lon and sector im interested in. (Sector 0 i have put to 0 in the ne2windia.nc file) Time is divided into 180, from 2000.2014. Im interested in all these years, so from time 0-180.
The NOx-em-anthro_input4MIPs_emissions_CIP_CEDS-2017-05-18_gn_200001-201412.nc is to large to send, but as you can see in my earlier post is it quite the same at ne2windia.nc, but global and with nonzero and zero values considering NOx_em_anthro variable, and with nonzero values considering the different sectors.
| ne2windia.nc (9.07 MB) ne2windia.nc |
RE: segmentation fault - Added by Hanne Byre over 6 years ago
I divided the large dataset into smaller, so her is a sample from the 13 first months.
| firstyear.nc (27.1 MB) firstyear.nc |
RE: segmentation fault - Added by Ralf Mueller over 6 years ago
Sorry, I misread your call and took setclonlatbox for sellonlatbox, sorry.
I checked the call and tracked the segfault down to the hdf5 library. Since you read in parallel from the same file (netcdf4 compressed), this can happen when hdf5 is installed non-thread-safe. Please use the option -L in your cdo call. I checked this with my latest-release version
% cdo -V
Climate Data Operators version 1.9.5 (http://mpimet.mpg.de/cdo)
System: x86_64-pc-linux-gnu
CXX Compiler: g++ -g -O3 -std=c++11 -Wall -fopenmp -march=native -ftree-vectorize -mavx2
CXX version : g++ (GCC) 8.2.1 20180831
C Compiler: gcc -g -O3 -std=gnu99 -Wall -fopenmp -march=native -ftree-vectorize -mavx2
C version : gcc (GCC) 8.2.1 20180831
F77 Compiler: gfortran -g -O2
F77 version : GNU Fortran (GCC) 8.2.1 20180831
Features: 15GB 8threads C++11 Fortran DATA PTHREADS OpenMP45 HDF5 NC4/HDF5 OPeNDAP SZ UDUNITS2 PROJ.4 XML2 MAGICS CURL FFTW3 AVX2
Libraries: HDF5/1.10.4 proj/5.2 xml2/2.9.8 curl/7.62.0
Filetypes: srv ext ieg grb1 grb2 nc1 nc2 nc4 nc4c nc5
CDI library version : 1.9.5
CGRIBEX library version : 1.9.1
GRIB_API library version : 2.7.3
NetCDF library version : 4.6.1 of Oct 23 2018 15:57:27 $
HDF5 library version : 1.10.4
EXSE library version : 1.4.0
FILE library version : 1.8.3
And it worked properly
cdo -L -v -add -setclonlatbox,0,7.5,37.5,67.5,97.5 firstyear.nc -setmisstoc,0 -enlargegrid,firstyear.nc ne2windia.nc nyfil.ncResults is uploaded (gziped).
hth
ralf
| nyfil.nc.gz (22.7 MB) nyfil.nc.gz |
RE: segmentation fault - Added by Hanne Byre over 6 years ago
Thank you so much! It worked if i skipped the -v in cdo -L -v -add -setclonlatbox,0,7.5,37.5,67.5,97.5 NOx-em-anthro_input4MIPs_emissions_CIP_CEDS-2017-05-18_gn_200001-201412.nc -setmisstoc,0 -enlargegrid,NOx-em-anthro_input4MIPs_emissions_CIP_CEDS-2017-05-18_gn_200001-201412.nc ne2windia.nc nyfil.nc
But, I wanted the values between lat: 0,7.5,37.5 and lon 67.5,97.5 to be zero in sector 0. That why I replaced the original values with zero and created the ne2windia.nc file (i have checked that the values in ne2windia.nc are 0)
Why isnt the values in nyfil.nc between lat: 0,7.5,37.5 and lon 67.5,97.5 sector:0 zero?
They are not zero in the file you generated either.
-Hanne
RE: segmentation fault - Added by Hanne Byre over 6 years ago
Is the -setclonlatbox,0,7.5,37.5,67.5,97.5 option not treating the sector since that is a own dim?
RE: segmentation fault - Added by Ralf Mueller over 6 years ago
IMO the setclonlatbox command works for your dataset. I tried this
cdo -f nc -setclonlatbox,100,7.5,37.5,67.5,97.5 -seltimestep,1 firstyear.nc fy_to_100.ncwith uploaded results. I took 100 instead of 0 because it's easier to find in a plot
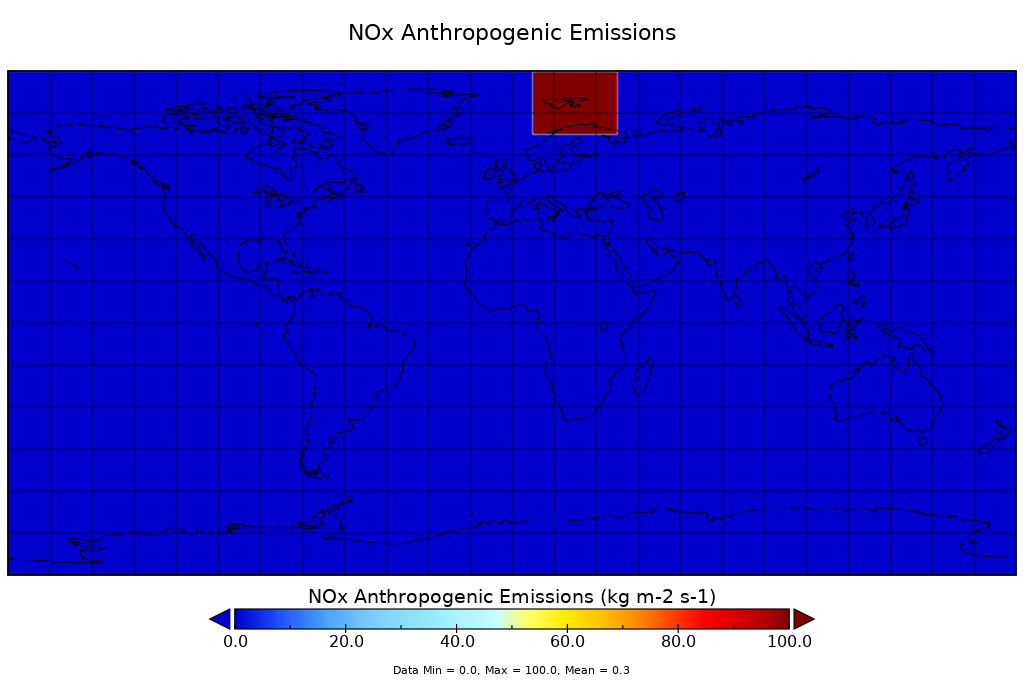
BTW: in my result files, the area has only zeros
cdo -infov -sellonlatbox,7.5,37.5,67.5,97.5 nyfil.nc
cdo(2) sellonlatbox: Process started
-1 : Date Time Level Gridsize Miss : Minimum Mean Maximum : Parameter name
1 : 2000-01-16 00:00:00 0 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
2 : 2000-01-16 00:00:00 1 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
3 : 2000-01-16 00:00:00 2 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
4 : 2000-01-16 00:00:00 3 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
5 : 2000-01-16 00:00:00 4 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
6 : 2000-01-16 00:00:00 5 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
7 : 2000-01-16 00:00:00 6 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
8 : 2000-01-16 00:00:00 7 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
9 : 2000-02-15 00:00:00 0 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
10 : 2000-02-15 00:00:00 1 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
11 : 2000-02-15 00:00:00 2 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
12 : 2000-02-15 00:00:00 3 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
13 : 2000-02-15 00:00:00 4 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
14 : 2000-02-15 00:00:00 5 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
15 : 2000-02-15 00:00:00 6 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
16 : 2000-02-15 00:00:00 7 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
17 : 2000-03-16 00:00:00 0 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
18 : 2000-03-16 00:00:00 1 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
19 : 2000-03-16 00:00:00 2 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
20 : 2000-03-16 00:00:00 3 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
21 : 2000-03-16 00:00:00 4 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
22 : 2000-03-16 00:00:00 5 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
23 : 2000-03-16 00:00:00 6 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
24 : 2000-03-16 00:00:00 7 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
25 : 2000-04-16 00:00:00 0 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
26 : 2000-04-16 00:00:00 1 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
27 : 2000-04-16 00:00:00 2 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
28 : 2000-04-16 00:00:00 3 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
29 : 2000-04-16 00:00:00 4 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
30 : 2000-04-16 00:00:00 5 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
31 : 2000-04-16 00:00:00 6 2700 0 : 0.0000 0.0000 0.0000 : NOx_em_anthro
....
RE: segmentation fault - Added by Hanne Byre over 6 years ago
Im so sorry, I did mix up the lon/lat, so i was looking at the wrong area.
Now everything works perfectly!
Thank you so much.
-Hanne
RE: segmentation fault - Added by Ralf Mueller over 6 years ago
you're welcome - good luck ... with whatever you do ;-)
RE: segmentation fault - Added by Hanne Byre over 6 years ago
One last question.. I want to do the same, but now my data looks like this:
dimensions:
lon = 720 ;
lat = 360 ;
time = 180 ;
bound = 2 ;
variables:
double lon(lon) ;
string lon:units = "degrees_east" ;
string lon:axis = "X" ;
string lon:standard_name = "longitude" ;
string lon:bounds = "lon_bnds" ;
string lon:realtopology = "circular" ;
lon:modulo = 360. ;
string lon:topology = "circular" ;
double lat(lat) ;
string lat:units = "degrees_north" ;
string lat:axis = "Y" ;
string lat:standard_name = "latitude" ;
string lat:bounds = "lat_bnds" ;
string lat:realtopology = "linear" ;
double time(time) ;
string time:units = "days since 1750-01-01 0:0:0" ;
string time:long_name = "time" ;
string time:standard_name = "time" ;
string time:calendar = "365_day" ;
string time:axis = "T" ;
string time:bounds = "time_bnds" ;
string time:realtopology = "linear" ;
double lon_bnds(lon, bound) ;
double lat_bnds(lat, bound) ;
float Gridcell\ area(lat, lon) ;
string Gridcell\ area:units = "m2" ;
float NOx_em_anthro_AGR(time, lat, lon) ;
string NOx_em_anthro_AGR:units = "kg m-2 s-1" ;
NOx_em_anthro_AGR:_FillValue = 1.e+20f ;
string NOx_em_anthro_AGR:cell_methods = "time: mean" ;
NOx_em_anthro_AGR:missing_value = 1.e+20f ;
string NOx_em_anthro_AGR:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_ENE(time, lat, lon) ;
string NOx_em_anthro_ENE:units = "kg m-2 s-1" ;
NOx_em_anthro_ENE:_FillValue = 1.e+20f ;
string NOx_em_anthro_ENE:cell_methods = "time: mean" ;
NOx_em_anthro_ENE:missing_value = 1.e+20f ;
string NOx_em_anthro_ENE:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_IND(time, lat, lon) ;
string NOx_em_anthro_IND:units = "kg m-2 s-1" ;
NOx_em_anthro_IND:_FillValue = 1.e+20f ;
string NOx_em_anthro_IND:cell_methods = "time: mean" ;
NOx_em_anthro_IND:missing_value = 1.e+20f ;
string NOx_em_anthro_IND:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_TRA(time, lat, lon) ;
string NOx_em_anthro_TRA:units = "kg m-2 s-1" ;
NOx_em_anthro_TRA:_FillValue = 1.e+20f ;
string NOx_em_anthro_TRA:cell_methods = "time: mean" ;
NOx_em_anthro_TRA:missing_value = 1.e+20f ;
string NOx_em_anthro_TRA:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_RCO(time, lat, lon) ;
string NOx_em_anthro_RCO:units = "kg m-2 s-1" ;
NOx_em_anthro_RCO:_FillValue = 1.e+20f ;
string NOx_em_anthro_RCO:cell_methods = "time: mean" ;
NOx_em_anthro_RCO:missing_value = 1.e+20f ;
string NOx_em_anthro_RCO:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_SOL(time, lat, lon) ;
string NOx_em_anthro_SOL:units = "kg m-2 s-1" ;
NOx_em_anthro_SOL:_FillValue = 1.e+20f ;
string NOx_em_anthro_SOL:cell_methods = "time: mean" ;
NOx_em_anthro_SOL:missing_value = 1.e+20f ;
string NOx_em_anthro_SOL:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_WAS(time, lat, lon) ;
string NOx_em_anthro_WAS:units = "kg m-2 s-1" ;
NOx_em_anthro_WAS:_FillValue = 1.e+20f ;
string NOx_em_anthro_WAS:cell_methods = "time: mean" ;
NOx_em_anthro_WAS:missing_value = 1.e+20f ;
string NOx_em_anthro_WAS:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_SHI(time, lat, lon) ;
string NOx_em_anthro_SHI:units = "kg m-2 s-1" ;
NOx_em_anthro_SHI:_FillValue = 1.e+20f ;
string NOx_em_anthro_SHI:cell_methods = "time: mean" ;
NOx_em_anthro_SHI:missing_value = 1.e+20f ;
string NOx_em_anthro_SHI:long_name = "NOx Anthropogenic Emissions" ;
// global attributes:
string :CICERO_title = "CEDS; sectors separated for CICERO usage" ;
string :CICERO_history = "idl script by A.S.Haslerud, CICERO" ;
string :CICERO_date = "Thu Aug 31 21:13:22 2017" ;
string :title = "Annual Anthropogenic Emissions of NOx prepared for input4MIPs" ;
string :source_id = "CEDS-2017-05-18" ;
string :references = "Hoesly, R. M., Smith, S. J., Feng, L., Klimont, Z., Janssens-Maenhout, G., Pitkanen, T., Seibert, J. J., Vu, L., Andres, R. J., Bolt, R. M., Bond, T. C., Dawidowski, L., Kholod, N., Kurokawa, J.-I., Li, M., Liu, L., Lu, Z., Moura, M. C. P., O\'Rourke, P. R., and Zhang, Q.: Historical (1750-2014) anthropogenic emissions of reactive gases and aerosols from the Community Emission Data System (CEDS), Geosci. Model Dev. Discuss., doi:10.5194/gmd-2017-43, in review, 2017." ;
string :dataset_version_number = "2017-05-18" ;
string :reporting_unit = "Mass flux of NOx, reported as NO2" ;
string :frequency = "mon" ;
and
dimensions:
time = 180 ;
lat = 60 ;
lon = 60 ;
bound = 2 ;
variables:
float NOx_em_anthro_AGR(time, lat, lon) ;
NOx_em_anthro_AGR:_FillValue = 1.e+20f ;
string NOx_em_anthro_AGR:cell_methods = "time: mean" ;
string NOx_em_anthro_AGR:long_name = "NOx Anthropogenic Emissions" ;
NOx_em_anthro_AGR:missing_value = 1.e+20f ;
string NOx_em_anthro_AGR:units = "kg m-2 s-1" ;
double lon(lon) ;
string lon:units = "degrees_east" ;
string lon:axis = "X" ;
string lon:standard_name = "longitude" ;
string lon:bounds = "lon_bnds" ;
string lon:realtopology = "circular" ;
lon:modulo = 360. ;
string lon:topology = "circular" ;
double lat(lat) ;
string lat:units = "degrees_north" ;
string lat:axis = "Y" ;
string lat:standard_name = "latitude" ;
string lat:bounds = "lat_bnds" ;
string lat:realtopology = "linear" ;
double time(time) ;
string time:units = "days since 1750-01-01 0:0:0" ;
string time:long_name = "time" ;
string time:standard_name = "time" ;
string time:calendar = "365_day" ;
string time:axis = "T" ;
string time:bounds = "time_bnds" ;
string time:realtopology = "linear" ;
double lon_bnds(lon, bound) ;
double lat_bnds(lat, bound) ;
float Gridcell\ area(lat, lon) ;
string Gridcell\ area:units = "m2" ;
float NOx_em_anthro_ENE(time, lat, lon) ;
string NOx_em_anthro_ENE:units = "kg m-2 s-1" ;
NOx_em_anthro_ENE:_FillValue = 1.e+20f ;
string NOx_em_anthro_ENE:cell_methods = "time: mean" ;
NOx_em_anthro_ENE:missing_value = 1.e+20f ;
string NOx_em_anthro_ENE:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_IND(time, lat, lon) ;
string NOx_em_anthro_IND:units = "kg m-2 s-1" ;
NOx_em_anthro_IND:_FillValue = 1.e+20f ;
string NOx_em_anthro_IND:cell_methods = "time: mean" ;
NOx_em_anthro_IND:missing_value = 1.e+20f ;
string NOx_em_anthro_IND:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_TRA(time, lat, lon) ;
string NOx_em_anthro_TRA:units = "kg m-2 s-1" ;
NOx_em_anthro_TRA:_FillValue = 1.e+20f ;
string NOx_em_anthro_TRA:cell_methods = "time: mean" ;
NOx_em_anthro_TRA:missing_value = 1.e+20f ;
string NOx_em_anthro_TRA:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_RCO(time, lat, lon) ;
string NOx_em_anthro_RCO:units = "kg m-2 s-1" ;
NOx_em_anthro_RCO:_FillValue = 1.e+20f ;
string NOx_em_anthro_RCO:cell_methods = "time: mean" ;
NOx_em_anthro_RCO:missing_value = 1.e+20f ;
string NOx_em_anthro_RCO:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_SOL(time, lat, lon) ;
string NOx_em_anthro_SOL:units = "kg m-2 s-1" ;
NOx_em_anthro_SOL:_FillValue = 1.e+20f ;
string NOx_em_anthro_SOL:cell_methods = "time: mean" ;
NOx_em_anthro_SOL:missing_value = 1.e+20f ;
string NOx_em_anthro_SOL:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_WAS(time, lat, lon) ;
string NOx_em_anthro_WAS:units = "kg m-2 s-1" ;
NOx_em_anthro_WAS:_FillValue = 1.e+20f ;
string NOx_em_anthro_WAS:cell_methods = "time: mean" ;
NOx_em_anthro_WAS:missing_value = 1.e+20f ;
string NOx_em_anthro_WAS:long_name = "NOx Anthropogenic Emissions" ;
float NOx_em_anthro_SHI(time, lat, lon) ;
string NOx_em_anthro_SHI:units = "kg m-2 s-1" ;
NOx_em_anthro_SHI:_FillValue = 1.e+20f ;
string NOx_em_anthro_SHI:cell_methods = "time: mean" ;
NOx_em_anthro_SHI:missing_value = 1.e+20f ;
string NOx_em_anthro_SHI:long_name = "NOx Anthropogenic Emissions" ;
// global attributes:
:_NCProperties = "version=1|netcdflibversion=4.4.1.1|hdf5libversion=1.10.0" ;
string :CICERO_title = "CEDS; sectors separated for CICERO usage" ;
string :CICERO_history = "idl script by A.S.Haslerud, CICERO" ;
string :CICERO_date = "Thu Aug 31 21:13:22 2017" ;
string :title = "Annual Anthropogenic Emissions of NOx prepared for input4MIPs" ;
string :source_id = "CEDS-2017-05-18" ;
string :references = "Hoesly, R. M., Smith, S. J., Feng, L., Klimont, Z., Janssens-Maenhout, G., Pitkanen, T., Seibert, J. J., Vu, L., Andres, R. J., Bolt, R. M., Bond, T. C., Dawidowski, L., Kholod, N., Kurokawa, J.-I., Li, M., Liu, L., Lu, Z., Moura, M. C. P., O\'Rourke, P. R., and Zhang, Q.: Historical (1750-2014) anthropogenic emissions of reactive gases and aerosols from the Community Emission Data System (CEDS), Geosci. Model Dev. Discuss., doi:10.5194/gmd-2017-43, in review, 2017." ;
string :dataset_version_number = "2017-05-18" ;
string :reporting_unit = "Mass flux of NOx, reported as NO2" ;
string :frequency = "mon" ;
:history = "Wed Nov 14 16:39:27 2018: ncap2 -s where(NOx_em_anthro_AGR>0.0000000000000000000000) NOx_em_anthro_AGR=0.00000000000000000000000; India.nc Newindia.nc\nWed Nov 14 16:34:42 2018: ncea -d lat,7.5,37.5 -d lon,67.5,97.5 NOx-em-anthro_input4MIPs_emissions_CMIP_CEDS-2017-05-18_gn_200001-201412_CICERO.nc India.nc" ;
:NCO = "4.6.9" ;
:nco_openmp_thread_number = 1 ;
}
I have to treat them as strings. How can I do that so that
cdo -L -v -add -setclonlatbox,0,67.5,97.5,7.5,37.5 NOx-em-anthro_input4MIPs_emissions_CMIP_CEDS-2017-05-18_gn_200001-201412_CICERO.nc -setmisstoc,0 -enlargegrid,NOx-em-anthro_input4MIPs_emissions_CMIP_CEDS-2017-05-18_gn_200001-201412_CICERO.nc Newindia.nc GlobIndia_agrzero.nc
works?
I get the error message "Error (cdf_get_att_text) : NetCDF: Attempt to convert between text & numbers"
-Hanne
RE: segmentation fault - Added by Ralf Mueller over 6 years ago
hi Hanne!
if possible, please upload some sample data. compression with gzip works good with netcdf.
i think the error comes from the data type used for attributes. your data seem to use strings instead of char arrays. this is not CF-compliant. but i have to have a closer look into the data to say more.
cheers
ralf
RE: segmentation fault - Added by Hanne Byre over 6 years ago
Hei!
I added the data (The global is from the 13 first months)
Thanks!
Cheers
Hanne
| first12M.nc (12.5 MB) first12M.nc | global | ||
| India.nc (3.04 MB) India.nc | the values are not put to zero here as in Newindia, but didnt have the Newindia file available here |
RE: segmentation fault - Added by Ralf Mueller over 6 years ago
hi Hanne!
I had no issues running
cdo -L -v -add -setclonlatbox,0,67.5,97.5,7.5,37.5 first12M.nc -setmisstoc,0 -enlargegrid,first12M.nc India.nc GlobIndia_agrzero.ncand uploaded the output. I suspect you are using and older CDO release than me (I used 1.9.5). If possible, please update your CDO installation to the current release.
Unfortunately subsequent runs of the same command ran into errors - this is very unstable, seems like a 50% change to avoid the error. The error again comed from within the hdf5 library.
sorry for the bad news
On another system with a thread-safe installation of hdf5 I get no errors at all - testes 100 runs without problems. So you should switch to a system with a threads-safe version of hdf5
hth
ralf
RE: segmentation fault - Added by Ralf Mueller over 6 years ago
hi again!
the Docker image with cdo-1.9.5 built-in runs without errors, too. Might be an easy way to use the latest release without manual installation. In case you are on MacOS, the macports port of CDO is well maintained and your call works, too.
hth
ralf
RE: segmentation fault - Added by Hanne Byre over 6 years ago
Thank you so much for the good news! I will try with the docker image. I tried to read this data with cdo version 1.6.4.
Hopefully it will work with the 1.9.5 version.
-Hanne