remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files
Added by Jeisson Javier Leal Rojas over 2 years ago
Hi all.
I am working in a cluster and I am trying to use remapnn, but I have not been able to do the remapping.
This is what I typed: cod remapnn tgt_grid.txt, input.nc output.nc
This is my tgt_grid.txt file:
#
# gridID 1
#
gridtype = lonlat
gridsize = 5040000
xname = LON
xlongname = longitude
xunits = degrees_east
yname = LAT
ylongname = latitude
yunits = degrees_north
xsize = 3600
ysize = 1400
xfirst = -179.95
xinc = 0.1
yfirst = 83.95
yinc = -0.1
This is the message I've got:
cdo (Abort): Unprocessed Input, could not process all Operators/Files
Unfortunately, this input.nc file is too heavy and I cannot upload it here. However, here there is a description of the file when using cdo sinful input.nc (With the link to download the file: http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc)
File format : NetCDF4
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Parameter name
1 : unknown git@git.mpimet c instant 1 1 20971520 1 F64 : lon_cell_centre
Grid coordinates :
1 : unstructured : points=20971520 nvertex=3
grid : number=15 position=1
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
clon : -3.141592 to 3.141593 radian
clat : -1.570211 to 1.570211 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
Vertical coordinates :
1 : surface : levels=1
cdo sinfon: Processed 1 variable [1.06s 1302MB].
[b382303@l40049 test_remap]$ ll
total 19292364
-rw-r--r-- 1 b382303 bb1153 18245294182 Mar 1 16:53 icon_grid.nc
-rw-r--r-- 1 b382303 bb1153 1610632480 Mar 1 19:17 selvar_clon.nc
-rw-r--r-- 1 b382303 bb1153 284 Mar 1 19:04 tgt_grid.grd
-rw-r--r-- 1 b382303 bb1153 283 Mar 1 17:44 tgt_grid.txt
[b382303@l40049 test_remap]$ lllf
bash: lllf: command not found
[b382303@l40049 test_remap]$ ls -alh
total 19G
drwxr-sr-x 2 b382303 bb1153 4.0K Mar 1 19:17 .
drwxr-sr-x 8 b382303 bb1153 4.0K Mar 1 19:09 ..
-rw-r--r-- 1 b382303 bb1153 17G Mar 1 16:53 icon_grid.nc
-rw-r--r-- 1 b382303 bb1153 1.6G Mar 1 19:17 selvar_clon.nc
-rw-r--r-- 1 b382303 bb1153 284 Mar 1 19:04 tgt_grid.grd
-rw-r--r-- 1 b382303 bb1153 283 Mar 1 17:44 tgt_grid.txt
[b382303@l40049 test_remap]$ cdo sinfon selvar_clon.nc
File format : NetCDF4
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Parameter name
1 : unknown git@git.mpimet c instant 1 1 20971520 1 F64 : lon_cell_centre
Grid coordinates :
1 : unstructured : points=20971520 nvertex=3
grid : number=15 position=1
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
clon : -3.141592 to 3.141593 radian
clat : -1.570211 to 1.570211 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
Vertical coordinates :
1 : surface : levels=1
cdo sinfon: Processed 1 variable [1.05s 1302MB].
[b382303@l40049 test_remap]$ cdo sinfon icon_grid.nc
File format : NetCDF4
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Parameter name
1 : unknown git@git.mpimet c instant 1 1 20971520 1 F64 : ifs2icon_cell_grid
2 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : ifs2icon_edge_grid
3 : unknown git@git.mpimet c instant 1 1 10485762 3 F64 : ifs2icon_vertex_grid
4 : unknown git@git.mpimet c instant 1 1 20971520 4 F64 : cell_area
5 : unknown git@git.mpimet c instant 1 1 10485762 3 F64 : dual_area
6 : unknown git@git.mpimet c instant 1 1 20971520 4 I32 : phys_cell_id
7 : unknown git@git.mpimet c instant 1 1 31457280 2 I32 : phys_edge_id
8 : unknown git@git.mpimet c instant 1 1 20971520 4 F64 : lon_cell_centre
9 : unknown git@git.mpimet c instant 1 1 20971520 4 F64 : lat_cell_centre
10 : unknown git@git.mpimet c instant 1 1 20971520 4 F64 : lat_cell_barycenter
11 : unknown git@git.mpimet c instant 1 1 20971520 4 F64 : lon_cell_barycenter
12 : unknown git@git.mpimet c instant 1 1 10485762 3 F64 : longitude_vertices
13 : unknown git@git.mpimet c instant 1 1 10485762 3 F64 : latitude_vertices
14 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : lon_edge_centre
15 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : lat_edge_centre
16 : unknown git@git.mpimet c instant 3 2 20971520 5 I32 : edge_of_cell
17 : unknown git@git.mpimet c instant 3 2 20971520 5 I32 : vertex_of_cell
18 : unknown git@git.mpimet c instant 2 3 31457280 6 I32 : adjacent_cell_of_edge
19 : unknown git@git.mpimet c instant 2 3 31457280 6 I32 : edge_vertices
20 : unknown git@git.mpimet c instant 6 4 10485762 7 I32 : cells_of_vertex
21 : unknown git@git.mpimet c instant 6 4 10485762 7 I32 : edges_of_vertex
22 : unknown git@git.mpimet c instant 6 4 10485762 7 I32 : vertices_of_vertex
23 : unknown git@git.mpimet c instant 1 1 20971520 4 F64 : cell_area_p
24 : unknown git@git.mpimet c instant 1 1 20971520 4 F64 : cell_elevation
25 : unknown git@git.mpimet c instant 1 1 20971520 4 I32 : cell_sea_land_mask
26 : unknown git@git.mpimet c instant 4 5 20971520 1 I32 : cell_domain_id
27 : unknown git@git.mpimet c instant 1 1 4 8 I32 : cell_no_of_domains
28 : unknown git@git.mpimet c instant 1 1 10485762 7 F64 : dual_area_p
29 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_length
30 : unknown git@git.mpimet c instant 2 3 31457280 6 F64 : edge_cell_distance
31 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : dual_edge_length
32 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edgequad_area
33 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_elevation
34 : unknown git@git.mpimet c instant 1 1 31457280 2 I32 : edge_sea_land_mask
35 : unknown git@git.mpimet c instant 2 3 31457280 6 F64 : edge_vert_distance
36 : unknown git@git.mpimet c instant 1 1 31457280 6 F64 : zonal_normal_primal_edge
37 : unknown git@git.mpimet c instant 1 1 31457280 6 F64 : meridional_normal_primal_edge
38 : unknown git@git.mpimet c instant 1 1 31457280 6 F64 : zonal_normal_dual_edge
39 : unknown git@git.mpimet c instant 1 1 31457280 6 F64 : meridional_normal_dual_edge
40 : unknown git@git.mpimet c instant 3 2 20971520 5 I32 : orientation_of_normal
41 : unknown git@git.mpimet c instant 1 1 20971520 5 I32 : cell_index
42 : unknown git@git.mpimet c instant 1 1 20971520 5 I32 : parent_cell_index
43 : unknown git@git.mpimet c instant 1 1 20971520 5 I32 : parent_cell_type
44 : unknown git@git.mpimet c instant 3 2 20971520 5 I32 : neighbor_cell_index
45 : unknown git@git.mpimet c instant 4 6 20971520 5 I32 : child_cell_index
46 : unknown git@git.mpimet c instant 1 1 20971520 5 I32 : child_cell_id
47 : unknown git@git.mpimet c instant 1 1 31457280 6 I32 : edge_index
48 : unknown git@git.mpimet c instant 1 1 31457280 6 I32 : edge_parent_type
49 : unknown git@git.mpimet c instant 1 1 10485762 7 I32 : vertex_index
50 : unknown git@git.mpimet c instant 6 4 10485762 7 I32 : edge_orientation
51 : unknown git@git.mpimet c instant 1 1 31457280 2 I32 : edge_system_orientation
52 : unknown git@git.mpimet c instant 1 1 20971520 5 I32 : refin_c_ctrl
53 : unknown git@git.mpimet c instant 1 1 28 9 I32 : index_c_list
54 : unknown git@git.mpimet c instant 1 1 14 10 I32 : start_idx_c
55 : unknown git@git.mpimet c instant 1 1 14 10 I32 : end_idx_c
56 : unknown git@git.mpimet c instant 1 1 31457280 6 I32 : refin_e_ctrl
57 : unknown git@git.mpimet c instant 1 1 48 11 I32 : index_e_list
58 : unknown git@git.mpimet c instant 1 1 24 12 I32 : start_idx_e
59 : unknown git@git.mpimet c instant 1 1 24 12 I32 : end_idx_e
60 : unknown git@git.mpimet c instant 1 1 10485762 7 I32 : refin_v_ctrl
61 : unknown git@git.mpimet c instant 1 1 26 13 I32 : index_v_list
62 : unknown git@git.mpimet c instant 1 1 13 14 I32 : start_idx_v
63 : unknown git@git.mpimet c instant 1 1 13 14 I32 : end_idx_v
64 : unknown git@git.mpimet c instant 1 1 31457280 6 I32 : parent_edge_index
65 : unknown git@git.mpimet c instant 4 6 31457280 6 I32 : child_edge_index
66 : unknown git@git.mpimet c instant 1 1 31457280 6 I32 : child_edge_id
67 : unknown git@git.mpimet c instant 1 1 10485762 7 I32 : parent_vertex_index
68 : unknown git@git.mpimet c instant 1 1 10485762 3 F64 : cartesian_x_vertices
69 : unknown git@git.mpimet c instant 1 1 10485762 3 F64 : cartesian_y_vertices
70 : unknown git@git.mpimet c instant 1 1 10485762 3 F64 : cartesian_z_vertices
71 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_middle_cartesian_x
72 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_middle_cartesian_y
73 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_middle_cartesian_z
74 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_dual_middle_cartesian_x
75 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_dual_middle_cartesian_y
76 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_dual_middle_cartesian_z
77 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_primal_normal_cartesian_x
78 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_primal_normal_cartesian_y
79 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_primal_normal_cartesian_z
80 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_dual_normal_cartesian_x
81 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_dual_normal_cartesian_y
82 : unknown git@git.mpimet c instant 1 1 31457280 2 F64 : edge_dual_normal_cartesian_z
83 : unknown git@git.mpimet c instant 1 1 20971520 1 F64 : cell_circumcenter_cartesian_x
84 : unknown git@git.mpimet c instant 1 1 20971520 1 F64 : cell_circumcenter_cartesian_y
85 : unknown git@git.mpimet c instant 1 1 20971520 1 F64 : cell_circumcenter_cartesian_z
Grid coordinates :
1 : unstructured : points=20971520 nvertex=3
grid : number=15 position=0
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
clon : -3.141592 to 3.141593 radian
clat : -1.570211 to 1.570211 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
2 : unstructured : points=31457280 nvertex=4
grid : number=15 position=0
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
elon : -3.141592 to 3.141593 radian
elat : -1.570323 to 1.570323 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
3 : unstructured : points=10485762 nvertex=6
grid : number=15 position=0
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
vlon : -3.141592 to 3.141592 radian
vlat : -1.570796 to 1.570796 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
4 : unstructured : points=20971520 nvertex=3
grid : number=15 position=1
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
clon : -3.141592 to 3.141593 radian
clat : -1.570211 to 1.570211 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
5 : unstructured : points=20971520
grid : number=15 position=0
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
6 : unstructured : points=31457280
grid : number=15 position=0
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
7 : unstructured : points=10485762
grid : number=15 position=0
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
8 : generic : points=4
9 : generic : points=28 (14x2)
10 : generic : points=14 (14x1)
11 : generic : points=48 (24x2)
12 : generic : points=24 (24x1)
13 : generic : points=26 (13x2)
14 : generic : points=13 (13x1)
Vertical coordinates :
1 : surface : levels=1
2 : generic : levels=3
3 : generic : levels=2
4 : generic : levels=6
5 : generic : levels=4
6 : generic : levels=4
cdo sinfon: Processed 85 variables [4.86s 6106MB].
I also tried to use selvar just to get one variable: cdo selvar,clon_vertices ./selvar_clon.nc
and this is the cdo sinfon selvar_clon.nc
File format : NetCDF4
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Parameter name
1 : unknown git@git.mpimet c instant 1 1 20971520 1 F64 : lon_cell_centre
Grid coordinates :
1 : unstructured : points=20971520 nvertex=3
grid : number=15 position=1
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
clon : -3.141592 to 3.141593 radian
clat : -1.570211 to 1.570211 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
Vertical coordinates :
1 : surface : levels=1
cdo sinfon: Processed 1 variable [0.92s 1302MB].
By doing cod remapnn tgt_grid.txt, selvar_clon.nc output.nc I got the exact same message
cdo (Abort): Unprocessed Input, could not process all Operators/Files
Does anyone have any idea what can I do? maybe the file is too big? how can I approach this issue?
Regards,
Jeisson Leal.
Replies (13)
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Ralf Mueller over 2 years ago
hi!
try
cdo -P 16 remapnn,tgt_grid.txt input.nc output.nc
instead
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Jeisson Javier Leal Rojas over 2 years ago
It worked, but not with the original input.nc file. Thank you so much Ralf, really appreciate it! you rock 
However, I do still have some doubts.
My original input.nc file has a lot of Grid coordinates:
Grid coordinates :
1 : unstructured : points=20971520 nvertex=3
grid : number=15 position=0
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
clon : -3.141592 to 3.141593 radian
clat : -1.570211 to 1.570211 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
2 : unstructured : points=31457280 nvertex=4
grid : number=15 position=0
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
elon : -3.141592 to 3.141593 radian
elat : -1.570323 to 1.570323 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
3 : unstructured : points=10485762 nvertex=6
grid : number=15 position=0
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
vlon : -3.141592 to 3.141592 radian
vlat : -1.570796 to 1.570796 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
4 : unstructured : points=20971520 nvertex=3
grid : number=15 position=1
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
clon : -3.141592 to 3.141593 radian
clat : -1.570211 to 1.570211 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
So when I tried what you suggested I've got the same error:
cdo remapnn (Abort): Unsupported generic coordinates (Variable: cell_no_of_domains)!
Not sure though if that message was due to so many Grid coordinates
Later, I used another nc file I had created from input.nc by issuing cdo selvar.
cdo selvar,clon_vertices ./selvar_clon.nc
This new selvar_clon.nc file has just one Grid coordinate. Looking at selvar_clon.nc
File format : NetCDF4
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Parameter name
1 : unknown git@git.mpimet c instant 1 1 20971520 1 F64 : lon_cell_centre
Grid coordinates :
1 : unstructured : points=20971520 nvertex=3
grid : number=15 position=1
uri : http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
clon : -3.141592 to 3.141593 radian
clat : -1.570211 to 1.570211 radian
available : cellbounds
uuid : 0f1e7d66-637e-11e8-913b-51232bb4d8f9
Vertical coordinates :
1 : surface : levels=1
cdo sinfon: Processed 1 variable [0.91s 1302MB].
With this new selvar_clon.nc and using cdo -P 16 remapnn,tgt_grid.txt selvar_clon.nc output.nc I got this outpu.nc file:
File format : NetCDF4
-1 : Institut Source T Steptype Levels Num Points Num Dtype : Parameter name
1 : unknown git@git.mpimet c instant 1 1 5040000 1 F64 : lon_cell_centre
Grid coordinates :
1 : lonlat : points=5040000 (3600x1400)
LON : -179.95 to 179.95 by 0.1 degrees_east circular
LAT : 83.95 to -55.95 by -0.1 degrees_north
Vertical coordinates :
1 : surface : levels=1
cdo sinfon: Processed 1 variable [0.02s 32MB].
And this output.nc file looks like what I needed!
However, as I said before. I still have some questions.
Can you please briefly explain me what my issue was and what -P 16 exactly does? and maybe also why it didn't work with the original input.nc file?
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Ralf Mueller over 2 years ago
Hi!
Is your input.nc an icon grid file maybe?
cdo selvar,clon_vertices ./selvar_clon.nc
- this is not a valid cdo call
- this does not work, because for CDO clon_vertices is not a data variable
Please provide a full output of
ncdump -h input.ncand let me know which data variable(s) of it you want to interpolation on the target lonlat grid.
cheers
ralf
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Jeisson Javier Leal Rojas over 2 years ago
Hi
Yes, my input.nc file is an ICON (Icosahedral Nonhydrostatic Weather and Climate Model) grid file provided in Levante cluster:
horizontal grid atm: /pool/data/ICON/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc
Here is the information when doing ncdump -h input.nc:
netcdf icon_grid {
dimensions:
cell = 20971520 ;
vertex = 10485762 ;
edge = 31457280 ;
nc = 2 ;
nv = 3 ;
ne = 6 ;
no = 4 ;
two_grf = 2 ;
max_chdom = 1 ;
cell_grf = 14 ;
edge_grf = 24 ;
vert_grf = 13 ;
max_stored_decompositions = 4 ;
variables:
double clon(cell) ;
clon:long_name = "center longitude" ;
clon:units = "radian" ;
clon:standard_name = "grid_longitude" ;
clon:bounds = "clon_vertices" ;
double clat(cell) ;
clat:long_name = "center latitude" ;
clat:units = "radian" ;
clat:standard_name = "grid_latitude" ;
clat:bounds = "clat_vertices" ;
double clon_vertices(cell, nv) ;
clon_vertices:units = "radian" ;
double clat_vertices(cell, nv) ;
clat_vertices:units = "radian" ;
double vlon(vertex) ;
vlon:long_name = "vertex longitude" ;
vlon:units = "radian" ;
vlon:standard_name = "grid_longitude" ;
vlon:bounds = "vlon_vertices" ;
double vlat(vertex) ;
vlat:long_name = "vertex latitude" ;
vlat:units = "radian" ;
vlat:standard_name = "grid_latitude" ;
vlat:bounds = "vlat_vertices" ;
double vlon_vertices(vertex, ne) ;
vlon_vertices:units = "radian" ;
double vlat_vertices(vertex, ne) ;
vlat_vertices:units = "radian" ;
double elon(edge) ;
elon:long_name = "edge midpoint longitude" ;
elon:units = "radian" ;
elon:standard_name = "grid_longitude" ;
elon:bounds = "elon_vertices" ;
double elat(edge) ;
elat:long_name = "edge midpoint latitude" ;
elat:units = "radian" ;
elat:standard_name = "grid_latitude" ;
elat:bounds = "elat_vertices" ;
double elon_vertices(edge, no) ;
elon_vertices:units = "radian" ;
double elat_vertices(edge, no) ;
elat_vertices:units = "radian" ;
double ifs2icon_cell_grid(cell) ;
ifs2icon_cell_grid:long_name = "ifs to icon cells" ;
ifs2icon_cell_grid:coordinates = "clon clat" ;
double ifs2icon_edge_grid(edge) ;
ifs2icon_edge_grid:long_name = "ifs to icon edge" ;
ifs2icon_edge_grid:coordinates = "elon elat" ;
double ifs2icon_vertex_grid(vertex) ;
ifs2icon_vertex_grid:long_name = "ifs to icon vertex" ;
ifs2icon_vertex_grid:coordinates = "vlon vlat" ;
double cell_area(cell) ;
cell_area:long_name = "area of grid cell" ;
cell_area:units = "m2" ;
cell_area:standard_name = "area" ;
cell_area:grid_type = "unstructured" ;
cell_area:coordinates = "clon clat" ;
cell_area:number_of_grid_in_reference = 1 ;
double dual_area(vertex) ;
dual_area:long_name = "areas of dual hexagonal/pentagonal cells" ;
dual_area:units = "m2" ;
dual_area:standard_name = "area" ;
dual_area:coordinates = "vlon vlat" ;
int phys_cell_id(cell) ;
phys_cell_id:long_name = "physical domain ID of cell" ;
phys_cell_id:grid_type = "unstructured" ;
phys_cell_id:coordinates = "clon clat" ;
phys_cell_id:number_of_grid_in_reference = 1 ;
int phys_edge_id(edge) ;
phys_edge_id:long_name = "physical domain ID of edge" ;
phys_edge_id:coordinates = "elon elat" ;
double lon_cell_centre(cell) ;
lon_cell_centre:long_name = "longitude of cell centre" ;
lon_cell_centre:units = "radian" ;
lon_cell_centre:grid_type = "unstructured" ;
lon_cell_centre:coordinates = "clon clat" ;
lon_cell_centre:number_of_grid_in_reference = 1 ;
double lat_cell_centre(cell) ;
lat_cell_centre:long_name = "latitude of cell centre" ;
lat_cell_centre:units = "radian" ;
lat_cell_centre:grid_type = "unstructured" ;
lat_cell_centre:coordinates = "clon clat" ;
lat_cell_centre:number_of_grid_in_reference = 1 ;
double lat_cell_barycenter(cell) ;
lat_cell_barycenter:long_name = "latitude of cell barycenter" ;
lat_cell_barycenter:units = "radian" ;
lat_cell_barycenter:grid_type = "unstructured" ;
lat_cell_barycenter:coordinates = "clon clat" ;
lat_cell_barycenter:number_of_grid_in_reference = 1 ;
double lon_cell_barycenter(cell) ;
lon_cell_barycenter:long_name = "longitude of cell barycenter" ;
lon_cell_barycenter:units = "radian" ;
lon_cell_barycenter:grid_type = "unstructured" ;
lon_cell_barycenter:coordinates = "clon clat" ;
lon_cell_barycenter:number_of_grid_in_reference = 1 ;
double longitude_vertices(vertex) ;
longitude_vertices:long_name = "longitude of vertices" ;
longitude_vertices:units = "radian" ;
longitude_vertices:coordinates = "vlon vlat" ;
double latitude_vertices(vertex) ;
latitude_vertices:long_name = "latitude of vertices" ;
latitude_vertices:units = "radian" ;
latitude_vertices:coordinates = "vlon vlat" ;
double lon_edge_centre(edge) ;
lon_edge_centre:long_name = "longitudes of edge midpoints" ;
lon_edge_centre:units = "radian" ;
lon_edge_centre:coordinates = "elon elat" ;
double lat_edge_centre(edge) ;
lat_edge_centre:long_name = "latitudes of edge midpoints" ;
lat_edge_centre:units = "radian" ;
lat_edge_centre:coordinates = "elon elat" ;
int edge_of_cell(nv, cell) ;
edge_of_cell:long_name = "edges of each cell\000\000vertices " ;
int vertex_of_cell(nv, cell) ;
vertex_of_cell:long_name = "vertices of each cell\000\000\000cells ad" ;
int adjacent_cell_of_edge(nc, edge) ;
adjacent_cell_of_edge:long_name = "cells adjacent to each edge" ;
int edge_vertices(nc, edge) ;
edge_vertices:long_name = "vertices at the end of of each edge" ;
int cells_of_vertex(ne, vertex) ;
cells_of_vertex:long_name = "cells around each vertex" ;
int edges_of_vertex(ne, vertex) ;
edges_of_vertex:long_name = "edges around each vertex" ;
int vertices_of_vertex(ne, vertex) ;
vertices_of_vertex:long_name = "vertices around each vertex" ;
double cell_area_p(cell) ;
cell_area_p:long_name = "area of grid cell" ;
cell_area_p:units = "m2" ;
cell_area_p:grid_type = "unstructured" ;
cell_area_p:coordinates = "clon clat" ;
cell_area_p:number_of_grid_in_reference = 1 ;
double cell_elevation(cell) ;
cell_elevation:long_name = "elevation at the cell centers" ;
cell_elevation:units = "m" ;
cell_elevation:grid_type = "unstructured" ;
cell_elevation:coordinates = "clon clat" ;
cell_elevation:number_of_grid_in_reference = 1 ;
int cell_sea_land_mask(cell) ;
cell_sea_land_mask:long_name = "sea (-2 inner, -1 boundary) land (2 inner, 1 boundary) mask for the cell" ;
cell_sea_land_mask:units = "2,1,-1,-" ;
cell_sea_land_mask:grid_type = "unstructured" ;
cell_sea_land_mask:coordinates = "clon clat" ;
cell_sea_land_mask:number_of_grid_in_reference = 1 ;
int cell_domain_id(cell, max_stored_decompositions) ;
cell_domain_id:long_name = "cell domain id for decomposition" ;
cell_domain_id:coordinates = "clon clat" ;
int cell_no_of_domains(max_stored_decompositions) ;
cell_no_of_domains:long_name = "number of domains for each decomposition" ;
double dual_area_p(vertex) ;
dual_area_p:long_name = "areas of dual hexagonal/pentagonal cells" ;
dual_area_p:units = "m2" ;
double edge_length(edge) ;
edge_length:long_name = "lengths of edges of triangular cells" ;
edge_length:units = "m" ;
edge_length:coordinates = "elon elat" ;
double edge_cell_distance(nc, edge) ;
edge_cell_distance:long_name = "distances between edge midpoint and adjacent triangle midpoints" ;
edge_cell_distance:units = "m" ;
double dual_edge_length(edge) ;
dual_edge_length:long_name = "lengths of dual edges (distances between triangular cell circumcenters)" ;
dual_edge_length:units = "m" ;
dual_edge_length:coordinates = "elon elat" ;
double edgequad_area(edge) ;
edgequad_area:long_name = "area around the edge formed by the two adjacent triangles" ;
edgequad_area:units = "m2" ;
edgequad_area:coordinates = "elon elat" ;
double edge_elevation(edge) ;
edge_elevation:long_name = "elevation at the edge centers" ;
edge_elevation:units = "m" ;
edge_elevation:coordinates = "elon elat" ;
int edge_sea_land_mask(edge) ;
edge_sea_land_mask:long_name = "sea (-2 inner, -1 boundary) land (2 inner, 1 boundary) mask for the cell" ;
edge_sea_land_mask:units = "2,1,-1,-" ;
edge_sea_land_mask:coordinates = "elon elat" ;
double edge_vert_distance(nc, edge) ;
edge_vert_distance:long_name = "distances between edge midpoint and vertices of that edge" ;
edge_vert_distance:units = "m" ;
double zonal_normal_primal_edge(edge) ;
zonal_normal_primal_edge:long_name = "zonal component of normal to primal edge" ;
zonal_normal_primal_edge:units = "radian" ;
double meridional_normal_primal_edge(edge) ;
meridional_normal_primal_edge:long_name = "meridional component of normal to primal edge" ;
meridional_normal_primal_edge:units = "radian" ;
double zonal_normal_dual_edge(edge) ;
zonal_normal_dual_edge:long_name = "zonal component of normal to dual edge" ;
zonal_normal_dual_edge:units = "radian" ;
double meridional_normal_dual_edge(edge) ;
meridional_normal_dual_edge:long_name = "meridional component of normal to dual edge" ;
meridional_normal_dual_edge:units = "radian" ;
int orientation_of_normal(nv, cell) ;
orientation_of_normal:long_name = "orientations of normals to triangular cell edges" ;
int cell_index(cell) ;
cell_index:long_name = "cell index" ;
int parent_cell_index(cell) ;
parent_cell_index:long_name = "parent cell index" ;
int parent_cell_type(cell) ;
parent_cell_type:long_name = "parent cell type" ;
int neighbor_cell_index(nv, cell) ;
neighbor_cell_index:long_name = "cell neighbor index" ;
int child_cell_index(no, cell) ;
child_cell_index:long_name = "child cell index" ;
int child_cell_id(cell) ;
child_cell_id:long_name = "domain ID of child cell" ;
int edge_index(edge) ;
edge_index:long_name = "edge index" ;
int edge_parent_type(edge) ;
edge_parent_type:long_name = "edge paren" ;
int vertex_index(vertex) ;
vertex_index:long_name = "vertices index" ;
int edge_orientation(ne, vertex) ;
edge_orientation:long_name = "edge orientation" ;
int edge_system_orientation(edge) ;
edge_system_orientation:long_name = "edge system orientation" ;
edge_system_orientation:coordinates = "elon elat" ;
int refin_c_ctrl(cell) ;
refin_c_ctrl:long_name = "refinement control flag for cells" ;
int index_c_list(two_grf, cell_grf) ;
index_c_list:long_name = "list of start and end indices for each refinement control level for cells" ;
int start_idx_c(max_chdom, cell_grf) ;
start_idx_c:long_name = "list of start indices for each refinement control level for cells" ;
int end_idx_c(max_chdom, cell_grf) ;
end_idx_c:long_name = "list of end indices for each refinement control level for cells" ;
int refin_e_ctrl(edge) ;
refin_e_ctrl:long_name = "refinement control flag for edges" ;
int index_e_list(two_grf, edge_grf) ;
index_e_list:long_name = "list of start and end indices for each refinement control level for edges" ;
int start_idx_e(max_chdom, edge_grf) ;
start_idx_e:long_name = "list of start indices for each refinement control level for edges" ;
int end_idx_e(max_chdom, edge_grf) ;
end_idx_e:long_name = "list of end indices for each refinement control level for edges" ;
int refin_v_ctrl(vertex) ;
refin_v_ctrl:long_name = "refinement control flag for vertices" ;
int index_v_list(two_grf, vert_grf) ;
index_v_list:long_name = "list of start and end indices for each refinement control level for vertices" ;
int start_idx_v(max_chdom, vert_grf) ;
start_idx_v:long_name = "list of start indices for each refinement control level for vertices" ;
int end_idx_v(max_chdom, vert_grf) ;
end_idx_v:long_name = "list of end indices for each refinement control level for vertices" ;
int parent_edge_index(edge) ;
parent_edge_index:long_name = "parent edge index" ;
int child_edge_index(no, edge) ;
child_edge_index:long_name = "child edge index" ;
int child_edge_id(edge) ;
child_edge_id:long_name = "domain ID of child edge" ;
int parent_vertex_index(vertex) ;
parent_vertex_index:long_name = "parent vertex index" ;
double cartesian_x_vertices(vertex) ;
cartesian_x_vertices:long_name = "vertex cartesian coordinate x on unit sp" ;
cartesian_x_vertices:units = "meters" ;
cartesian_x_vertices:coordinates = "vlon vlat" ;
double cartesian_y_vertices(vertex) ;
cartesian_y_vertices:long_name = "vertex cartesian coordinate y on unit sp" ;
cartesian_y_vertices:units = "meters" ;
cartesian_y_vertices:coordinates = "vlon vlat" ;
double cartesian_z_vertices(vertex) ;
cartesian_z_vertices:long_name = "vertex cartesian coordinate z on unit sp" ;
cartesian_z_vertices:units = "meters" ;
cartesian_z_vertices:coordinates = "vlon vlat" ;
double edge_middle_cartesian_x(edge) ;
edge_middle_cartesian_x:long_name = "prime edge center cartesian coordinate x on unit sphere" ;
edge_middle_cartesian_x:units = "meters" ;
edge_middle_cartesian_x:coordinates = "elon elat" ;
double edge_middle_cartesian_y(edge) ;
edge_middle_cartesian_y:long_name = "prime edge center cartesian coordinate y on unit sphere" ;
edge_middle_cartesian_y:units = "meters" ;
edge_middle_cartesian_y:coordinates = "elon elat" ;
double edge_middle_cartesian_z(edge) ;
edge_middle_cartesian_z:long_name = "prime edge center cartesian coordinate z on unit sphere" ;
edge_middle_cartesian_z:units = "meters" ;
edge_middle_cartesian_z:coordinates = "elon elat" ;
double edge_dual_middle_cartesian_x(edge) ;
edge_dual_middle_cartesian_x:long_name = "dual edge center cartesian coordinate x on unit sphere" ;
edge_dual_middle_cartesian_x:units = "meters" ;
edge_dual_middle_cartesian_x:coordinates = "elon elat" ;
double edge_dual_middle_cartesian_y(edge) ;
edge_dual_middle_cartesian_y:long_name = "dual edge center cartesian coordinate y on unit sphere" ;
edge_dual_middle_cartesian_y:units = "meters" ;
edge_dual_middle_cartesian_y:coordinates = "elon elat" ;
double edge_dual_middle_cartesian_z(edge) ;
edge_dual_middle_cartesian_z:long_name = "dual edge center cartesian coordinate z on unit sphere" ;
edge_dual_middle_cartesian_z:units = "meters" ;
edge_dual_middle_cartesian_z:coordinates = "elon elat" ;
double edge_primal_normal_cartesian_x(edge) ;
edge_primal_normal_cartesian_x:long_name = "unit normal to the prime edge 3D vector, coordinate x" ;
edge_primal_normal_cartesian_x:units = "meters" ;
edge_primal_normal_cartesian_x:coordinates = "elon elat" ;
double edge_primal_normal_cartesian_y(edge) ;
edge_primal_normal_cartesian_y:long_name = "unit normal to the prime edge 3D vector, coordinate y" ;
edge_primal_normal_cartesian_y:units = "meters" ;
edge_primal_normal_cartesian_y:coordinates = "elon elat" ;
double edge_primal_normal_cartesian_z(edge) ;
edge_primal_normal_cartesian_z:long_name = "unit normal to the prime edge 3D vector, coordinate z" ;
edge_primal_normal_cartesian_z:units = "meters" ;
edge_primal_normal_cartesian_z:coordinates = "elon elat" ;
double edge_dual_normal_cartesian_x(edge) ;
edge_dual_normal_cartesian_x:long_name = "unit normal to the dual edge 3D vector, coordinate x" ;
edge_dual_normal_cartesian_x:units = "meters" ;
edge_dual_normal_cartesian_x:coordinates = "elon elat" ;
double edge_dual_normal_cartesian_y(edge) ;
edge_dual_normal_cartesian_y:long_name = "unit normal to the dual edge 3D vector, coordinate y" ;
edge_dual_normal_cartesian_y:units = "meters" ;
edge_dual_normal_cartesian_y:coordinates = "elon elat" ;
double edge_dual_normal_cartesian_z(edge) ;
edge_dual_normal_cartesian_z:long_name = "unit normal to the dual edge 3D vector, coordinate z" ;
edge_dual_normal_cartesian_z:units = "meters" ;
edge_dual_normal_cartesian_z:coordinates = "elon elat" ;
double cell_circumcenter_cartesian_x(cell) ;
cell_circumcenter_cartesian_x:long_name = "cartesian position of the prime cell circumcenter on the unit sphere, coordinate x" ;
cell_circumcenter_cartesian_x:units = "meters" ;
cell_circumcenter_cartesian_x:coordinates = "clon clat" ;
double cell_circumcenter_cartesian_y(cell) ;
cell_circumcenter_cartesian_y:long_name = "cartesian position of the prime cell circumcenter on the unit sphere, coordinate y" ;
cell_circumcenter_cartesian_y:units = "meters" ;
cell_circumcenter_cartesian_y:coordinates = "clon clat" ;
double cell_circumcenter_cartesian_z(cell) ;
cell_circumcenter_cartesian_z:long_name = "cartesian position of the prime cell circumcenter on the unit sphere, coordinate z" ;
cell_circumcenter_cartesian_z:units = "meters" ;
cell_circumcenter_cartesian_z:coordinates = "clon clat" ;
// global attributes:
:title = "ICON grid description" ;
:institution = "Max Planck Institute for Meteorology/Deutscher Wetterdienst" ;
:source = "git@git.mpimet.mpg.de:GridGenerator.git" ;
:revision = "d00fcac1f61fa16c686bfe51d1d8eddd09296cb5" ;
:date = "20180529 at 222250" ;
:user_name = "Rene Redler (m300083)" ;
:os_name = "Linux 2.6.32-696.18.7.el6.x86_64 x86_64" ;
:uuidOfHGrid = "0f1e7d66-637e-11e8-913b-51232bb4d8f9" ;
:grid_mapping_name = "lat_long_on_sphere" ;
:crs_id = "urn:ogc:def:cs:EPSG:6.0:6422" ;
:crs_name = "Spherical 2D Coordinate System" ;
:ellipsoid_name = "Sphere" ;
:semi_major_axis = 6371229. ;
:inverse_flattening = 0. ;
:grid_level = 9 ;
:grid_root = 2 ;
:grid_ID = 1 ;
:parent_grid_ID = 0 ;
:no_of_subgrids = 1 ;
:start_subgrid_id = 1 ;
:max_childdom = 1 ;
:boundary_depth_index = 0 ;
:rotation_vector = 0., 0., 0. ;
:grid_geometry = 1 ;
:grid_cell_type = 3 ;
:mean_edge_length = 7510.64679407352 ;
:mean_dual_edge_length = 4336.34434517703 ;
:mean_cell_area = 24323517.8092827 ;
:mean_dual_cell_area = 48647026.3398971 ;
:domain_length = 40031612.4414765 ;
:domain_height = 40031612.4414765 ;
:sphere_radius = 6371229. ;
:domain_cartesian_center = 0., 0., 0. ;
:centre = 252 ;
:rotation = "37deg around z-axis" ;
:coverage = "global" ;
:symmetry = "along equator" ;
:topography = "modified SRTM30" ;
:subcentre = 1 ;
:number_of_grid_used = 15 ;
:history = "Thu Aug 16 11:05:44 2018: ncatted -O -a ICON_grid_file_uri,global,m,c,http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc icon_grid_0015_R02B09_G.nc test.nc\nWed May 30 08:50:27 2018: ncatted -a centre,global,c,i,252 -a rotation,global,c,c,37deg around z-axis -a coverage,global,c,c,global -a symmetry,global,c,c,along equator -a topography,global,c,c,modified SRTM30 -a subcentre,global,c,i,1 -a number_of_grid_used,global,c,i,15 -a ICON_grid_file_uri,global,c,c,http://icon-downloads.mpimet.mpg.de/grids/public/icon_grid_0015_R02B09_G.nc Earth_Global_IcosSymmetric_4932m_rotatedZ37d_modified_srtm30_1min.nc icon_grid_0015_R02B09_G.nc\n/mnt/lustre01/work/mh0287/users/rene/GridGenerator/build/x86_64-unknown-linux-gnu/bin/grid_command" ;
:ICON_grid_file_uri = "http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc" ;
:NCO = "netCDF Operators version 4.7.5 (Homepage = http://nco.sf.net, Code = http://github.com/nco/nco)" ;
}
from that input.nc I want to get the Lon-lat coordinates. There are many Lon lat variables, but I was thinking to use double clon_vertices(cell, nv) and double clat_vertices(cell, nv) to do the interpolation with my target grid.
After finally getting this interpolated grid I plan to merge that grid information to a Precipitation and Temperature files measured every 30min from 10.02.2020 to 01.03.2022. I already have the precipitation and Temperature nc files, but they do not have the lon lat coordinates I need.
ncdump -h precipitation nc file:
netcdf pre_30min {
dimensions:
time = UNLIMITED ; // (36001 currently)
ncells = 20971520 ;
variables:
double time(time) ;
time:standard_name = "time" ;
time:units = "minutes since 2020-1-20 00:00:00" ;
time:calendar = "proleptic_gregorian" ;
time:axis = "T" ;
float pr(time, ncells) ;
pr:standard_name = "pr" ;
pr:long_name = "precipitation flux" ;
pr:units = "kg m-2 s-1" ;
pr:param = "52.1.0" ;
pr:CDI_grid_type = "unstructured" ;
pr:number_of_grid_in_reference = 1 ;
pr:level_type = "atmosphere" ;
// global attributes:
:CDI = "Climate Data Interface version 2.0.4 (https://mpimet.mpg.de/cdi)" ;
:Conventions = "CF-1.6" ;
:number_of_grid_used = 15 ;
:grid_file_uri = "http://icon-downloads.mpimet.mpg.de/grids/public/mpim/0015/icon_grid_0015_R02B09_G.nc" ;
:uuidOfHGrid = "0f1e7d66-637e-11e8-913b-51232bb4d8f9" ;
:source = "git@gitlab.dkrz.de:icon/icon-aes.git@87a1eaded69e87889053ec3ee079b895c4e7bad8" ;
:institution = "Max Planck Institute for Meteorology/Deutscher Wetterdienst" ;
:title = "ICON simulation" ;
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Ralf Mueller over 2 years ago
Jeisson Javier Leal Rojas wrote in RE: remapnn in CDO - cdo (Abort): Unprocessed Input, coul...:
Hi
Yes, my input.nc file is an ICON (Icosahedral Nonhydrostatic Weather and Climate Model) grid file provided in Levante cluster:
That was my suspicion. IMO that does not make sense
Here is the information when doing ncdump -h input.nc:
[...]
from that input.nc I want to get the Lon-lat coordinates. There are many Lon lat variables, but I was thinking to use double clon_vertices(cell, nv) and double clat_vertices(cell, nv) to do the interpolation with my target grid.
What do you want to do with the coordinates? horizontal interpolation of data from one grid to another does not need the interpolation of the coordinates. you already have your new coordinates in the target grid.
So please, explain, why you think you need to perform an interpolation ONTO the source coordinates?
After finally getting this interpolated grid I plan to merge that grid information to a Precipitation and Temperature files measured every 30min from 10.02.2020 to 01.03.2022. I already have the precipitation and Temperature nc files, but they do not have the lon lat coordinates I need.
No offense, you seem to confuse things regarding horizontal interpolation. Here is how I see it
- you need to have source and target coordinates of your data variables
- which you already have: targetGrid.txt and the icon grid file with the cell coordinates
- CF-conform netcdf files declare the coordinates of data variables ether by dimension and corresponding coordinate variable names (for lonlat grids) or by using the
coordinatesattribute of a data variable (in the case of ICON)
- depending on the interpolation method, CDO computes interpolation weights for the given source and target grid
- then these weights are applied in a matrix-vector operation on the source data
- the result us the data variable on the target grid
Hence it is absolutely not needed to perform an interpolation on coordinates. you already know the target coordinates!
in case the input data and grids are CF-conform, CDO will figure out the coordinates and bounds (if needed) of do all the above steps in one call like remapnn
ncdump -h precipitation nc file:
[...]
you see in that last block it is obvious:
float pr(time, ncells) ;
pr:standard_name = "pr" ;
pr:long_name = "precipitation flux" ;
pr:units = "kg m-2 s-1" ;
pr:param = "52.1.0" ;
pr:CDI_grid_type = "unstructured" ;
pr:number_of_grid_in_reference = 1 ;
pr:level_type = "atmosphere" ;
this is not a CF-conform description, because the coordinates attribute is not used. At least it is somewhat reasonable, because the coordinates are not store in the same file as the data. At the end if make your life harder, because you have to find the right grid on your own. You can workaround this with the setgrid operator like
cdo -remapnn,targetGrid.txt -setgrid,icon_grid.nc icon_model_data.nc lonlat_output.nc
If the cell grid is not the first grid in the icon grid file (sometimes that's the case), you select the grid by padding ':2' or ':3' at the
icon_grid.nc filename
hth
ralf
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Jeisson Javier Leal Rojas over 2 years ago
No problem, I know I am new with CDO.
This is what I did in the end:
1) I created a tgt_grid.txt file from a nc file different from ICON. The idea is to regrid the ICON data to the tgt_grid.txt I've got.
2) I applied you suggestion with the ICON grid file, but I used gennn instead of remapnn:
cdo -P 16 gennn,tgt_grid.txt input.nc nn_weights.nc
3) I used remap of the precipitation data given in ICON:
cdo remap,tgt_grid.txt,nn_weights.nc pre_30min.nc pre_output_30min.nc
I got this nc file in the end, which was what I was looking for:
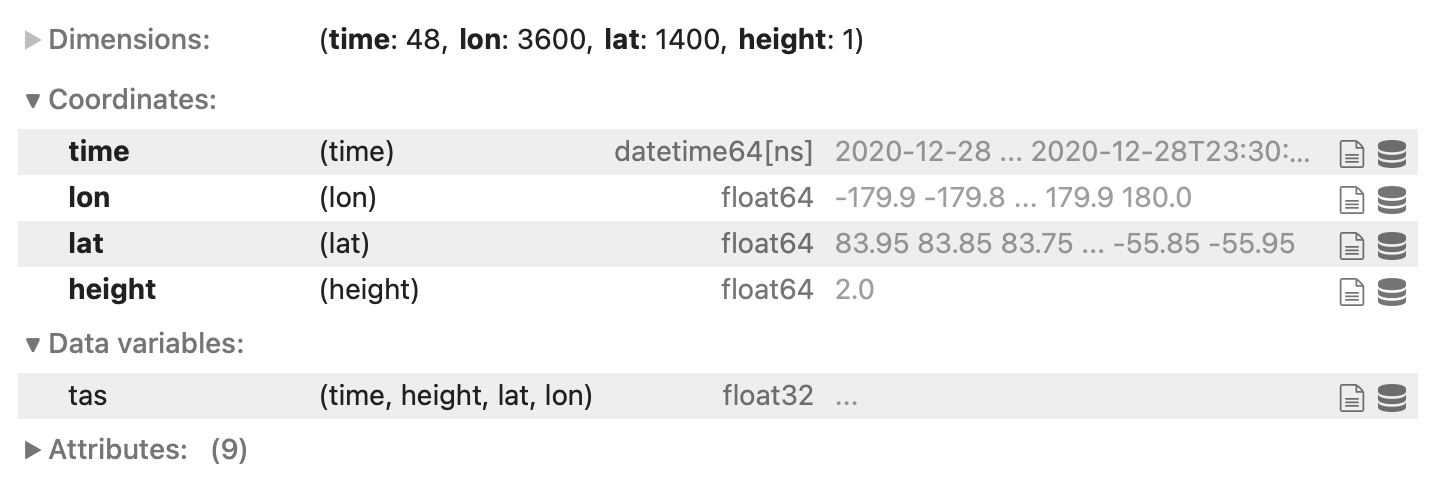
this picture is for the temperature, but I've got the same for precipitation.
Now my precipitation and temperature data have the. coordinates I was looking for after regridding.
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Ralf Mueller over 2 years ago
Jeisson Javier Leal Rojas wrote in RE: remapnn in CDO - cdo (Abort): Unprocessed Input, coul...:
No problem, I know I am new with CDO.
all cool. sorry if I was rude or so - I didn't mean to
This is what I did in the end:
1) I created a tgt_grid.txt file from a nc file different from ICON. The idea is to regrid the ICON data to the tgt_grid.txt I've got.
2) I applied you suggestion with the ICON grid file, but I used gennn instead of remapnn:
[...]3) I used remap of the precipitation data given in ICON:
[...]
Perfect!
I got this nc file in the end, which was what I was looking for:
this picture is for the temperature, but I've got the same for precipitation.
Now my precipitation and temperature data have the. coordinates I was looking for after regridding.
Nice job
maybe some things to note for the future:- ICON output may have data in 3 different grids: cells, edges and vertices. in general the variables on edges use useless, because it is the scalar result of the real vector and the edge-normal vector.
- the order of variables in the file determines the order if the grids. usually cell grid comes first, but not in every grid files. It depends on the grid generator, that created that specific grid file
So lets say you have model output on different grids, but you only want to interpolate the variables on cells. you can select all of them by their name of by their grid id.
cdo sinfov <icon_input>will show you table with variables and the corresponding grid in the icon model output. Lets say this id = 3. the you can to the above call like this
cdo -P8 -gennn,target_grid.txt -setgrid,icon_grid.nc:2 -selgrid,3 icon_input.nc
I don;t think you will encounter such situation in the real world, but I wanted to illustrate the different options. in many cased cell variables/grids are at first position anyway.
have a good weekend!
cheers
ralf
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Lokahith Agasthya about 1 year ago
Hi Jeisson Javier,
I am facing the same problem with -gennn as you while working with an ICON data file. Can you please share the tgt_grid.txt file you finally used or tell me how you generated it to solve the problem?
Thanks
Lokahith
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Jeisson Javier Leal Rojas about 1 year ago
Hi Lokahith,
If you scroll all the way up to the top, you will see the tgt_grid.txt content. I will just put it here once again 
# # gridID 1 # gridtype = lonlat gridsize = 5040000 xname = LON xlongname = longitude xunits = degrees_east yname = LAT ylongname = latitude yunits = degrees_north xsize = 3600 ysize = 1400 xfirst = -179.95 xinc = 0.1 yfirst = 83.95 yinc = -0.1
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Jeisson Javier Leal Rojas about 1 year ago
Or
# # gridID 1 # gridtype = lonlat gridsize = 5040000 xname = lon xlongname = longitude xunits = degrees_east yname = lat ylongname = latitude yunits = degrees_north xsize = 3600 ysize = 1400 xfirst = -179.95 xinc = 0.1 yfirst = 83.95 yinc = -0.1
it all depends on the input file and how you want do the remapping
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Lokahith Agasthya about 1 year ago
Hi Jeisson Javier,
Thanks a lot! Unfortunately that doesn't fix the issue for me - I was already using this format.
I get the error gennn (Abort): Unsupported generic coordinates (Variable: cell_no_of_domains)!
I have 2 differences from your case -
a) I am using the R2B10 grid file for ICON, which is the 2.5 km resolution grid. Maybe this causes a major difference because this doesn't have any information on the vertical grid. The vertical grid information is saved on another *vgrid.nc file.
b) My target grid doesn't cover the entire globe - it only covers a region of my interest from 5 to 35 N and 65 to 90 E.
I don't see why point b) should be an issue for nearest neighbour interpolation. Maybe it is to do with point a) ?
gridtype = lonlat
gridsize = 1200000
xsize = 1200
ysize = 1000
xname = lon
xlongname = longitude
xunits = degrees_east
yname = lat
ylongname = latitude
yunits = degrees_north
xfirst = 65.00
xinc = 0.025
yfirst = 5.00
yinc = 0.025
I appreciate any help/hints.
Thanks,
Lokahith
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Jeisson Javier Leal Rojas about 1 year ago
Hi
can you share your nc file? or a sample of it?
RE: remapnn in CDO - cdo (Abort): Unprocessed Input, could not process all Operators/Files - Added by Lokahith Agasthya about 1 year ago
Hi Jeisson,
The nc file is quite massive. You can find it in the same folder as your grid -
/pool/data/ICON/grids/public/mpim/0017/icon_grid_0017_R02B10_G.nc
PS - I actually managed to find a solution. I just had to add -selgrid,1 after the grid description txt file. I have not checked the weights file and the created interpolated data yet to see if everything worked well, but at least it executed!
Thanks a lot for your help. I will write again if I face any problems.
Lokahith
