Source cell masking in calculating remap weights.
Added by Brendan DeTracey over 4 years ago
Is is possible to specify source grid masking calculating remapping weights? This would be like the imask variable in a SCRIP style source grid definition. This would allow users to remap grids that have duplicate vertices for some cells such as the output from many current ocean models(tripolar grids). Currently, CDO may not interpolate these grids because they fail grid verification, but the grid points that fail verification are (usually) NaN and should not be used in weight calculations. Either a source grid mask option, and/or a genweight option to ignore all cells with duplicate vertices(with a warning message) would be extremely useful. And it would reduce the number of questions from NEMO users! If this is possible should I create an Issue for the suggestion?
Replies (29)
RE: Source cell masking in calculating remap weights. - Added by Ralf Mueller over 4 years ago
Happy New Year, Brendan!
Reduce the number of questions - that's like the wisdom of the year ;-)
I have to admit, that I did not fully understand your question: Do you have missing values in the coordinates?
An example file might by helpful
cheers
ralf
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
- Add an option to weight generation that uses
verifygridto generate this mask of rubbish points and apply it automatically. - Add an option to weight generation to apply an arbitrary mask provided by the user.
- Add an option to
verifygridto write this mask to a file.
A less ideal but simpler solution would be to add an option to weight generation that simply ignores cells that it detects are invalid, and prints a warning.
As I searched for an example I found an even more pathological example. Try running a verbose verifygrid on it. The output is too long to paste here.
$ cdo gencon,global_1 zos_Omon_CNRM-CM6-1_historical_r1i1p1f2_gn_185008.nc test.nc cdo gencon: YAC first order conservative weights from curvilinear (362x294) to lonlat (360x180) grid, with source mask (65087) cdo gencon: 5%ERROR: invalid cell Aborting in file clipping.c, line 1295 ...
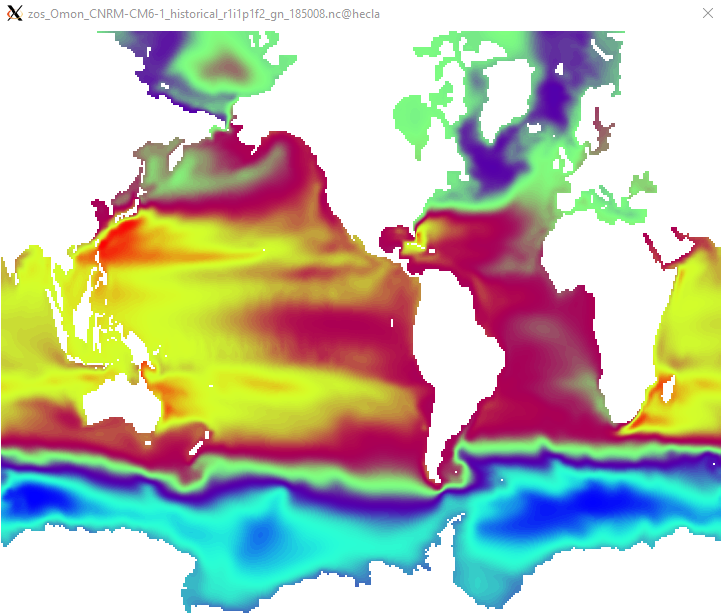
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
For my case, I am talking about conservative weights. Bilinear weights do not fail on this file.
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
Ack. They do not fail, but they would still benefit from rubbish cell masking.
$ cdo remapbil,global_1 zos_Omon_CNRM-CM6-1_historical_r1i1p1f2_gn_185008.nc test.nc cdo remapbil: Bilinear weights from curvilinear (362x294) to lonlat (360x180) grid, with source mask (65087) cdo remapbil: Processed 106428 values from 1 variable over 1 timestep [0.14s 50MB].<pre> !Screenshot%202021-01-07%20120016.png!
RE: Source cell masking in calculating remap weights. - Added by Ralf Mueller over 4 years ago
I am too familiar with the details of tripolar grid, but MPIOM (the ocean model developed here at MPIMET) can be run on a tripolar grid, too. Depending the differences in the description and the exact layout of it CDO might already have a reasonable workaround for you.
Your suggestion regarding the possible output of a mask by varifygrid sounds like a good approach. When I run it on your input, it gives:
cdo verifygrid: Grid consists of 106428 (362x294) cells (type: curvilinear), of which cdo verifygrid: 30 cells have 3 vertices cdo verifygrid: 106036 cells have 4 vertices cdo verifygrid: 30 cells have duplicate vertices cdo verifygrid: 362 cells have unusable vertices cdo verifygrid: 947 cells are not unique cdo verifygrid: 369 cells are non-convex cdo verifygrid: 9 cells have their vertices arranged in a clockwise order cdo verifygrid: 153 cells have their center point located outside their boundaries cdo verifygrid: lon : -179.9965 to 179.9903 degrees cdo verifygrid: lat : -79.00794 to 89.74177 degrees cdo verifygrid: Processed 1 variable [0.17s 62MB].
So the first question is: Does this operator really identify the correct cells to be taken into account as 'valid' ones? Are the 106036 cells the good ones and the all other cells are bad? I guess you want to keep the curvilinear structure of the file intact.
I will talk to Uwe about this and he probably has more (and more sophisticated) questions ;-)
best wishes
ralf
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
- have less than three vertices
- are not unique
- have their vertices arranged in a clockwise order
- have their center point located outside their boundaries
The only hitch for unstructured grids is a cell may have less than the maximum possible number of vertices. The excess vertices are dealt with by continuing to wind around the cell counterclockwise.
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
I really wish there was an edit button...I meant to be talking about curvilinear grids, not unstructured.
For curvilinear grids change 1 above to:- have less than four vertices
and add:
5. are non-convex
I think the four tests still apply to unstructured grids. Unstructured grid cell need to be allowed to be non-convex.
RE: Source cell masking in calculating remap weights. - Added by Uwe Schulzweida over 4 years ago
The source grid mask is automatically generated from the missing values of the input data. The coordinates are already processed before. If some coordinates cannot be processed, they must be removed beforehand:
cdo remapcon,global_1 -selindexbox,2,361,2,293 zos_Omon_CNRM-CM6-1_historical_r1i1p1f2_gn_185008.nc resultIf the source grid mask is constant, you can extract this mask and use reducegrid to remove all unused cells:
cdo setmisstoc,0 -gtc,-1000 zos_Omon_CNRM-CM6-1_historical_r1i1p1f2_gn_185008.nc mask cdo remapcon,global_1 -reducegrid,mask zos_Omon_CNRM-CM6-1_historical_r1i1p1f2_gn_185008.nc result
Cheers,
Uwe
RE: Source cell masking in calculating remap weights. - Added by Ralf Mueller over 4 years ago
hi Uwe!
I had excluded the reducegrid operator from my (mental) options for a workaround, because it creates an unstructured grid. But as an intermediate step towards another target grid, the unstructured grid is fine - wonderful solution in my opinion.
here is some more info about reducegrid
cheers
ralf
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
- run
cdo -v verifygrid - write a script to parse stdout to get indices for rubbish cells, format them to a comma delimited string and save the string to a text file
- pass the indices string from the text files to
cdo gencon,global_1 -delgridcell,indices
The proper(in both english and french sense) solution would be allowing options to verifygrid such that it generates a mask file. The mask would be determined by letting the user choose which tests a cell fails in verifygrid.
When I choose to use cdo, I do it as a personal choice to minimize code writing. I will hack up my own bash, awk, or python solution to this and it will not take very long. However, you will continue to have users asking why their CMIP ocean model data will not remap in cdo. 
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
I'll provide real examples of what I mean, whenever I manage to restore my VPN connection to work.
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
Here is the demo. I suppose my goal is trying to get cdo to remapcon CMIP6 ocean model output, much of which is on curvilinear tripolar grids. I do not understand why it will not work if the grid is reduced from curvilinear to unstructured. The problem is that much of the CMIP6 ocean model output is improperly formatted. Sometimes the grid vertices have been filled with fake values, and these fake values break remapcon.
The demo is my imperfect attempt to get remapcon to work on all the CMIP6 ocean grids. One problem is that I am throwing away all duplicates, but I want to throw away only duplicates with a data value of NaN. I can't think of a solution within cdo.
| remapcon_demo.tar.gz (7.64 MB) remapcon_demo.tar.gz |
RE: Source cell masking in calculating remap weights. - Added by Ralf Mueller over 4 years ago
Hi Brendan!
please have a look into the uploaded script. It's basically Uwe's suggestion working on your example inputs. Verifygrid shows only a single warning on one of the final output files. Can you comment on what exactly is wrong with the output from your point of view
I could not find NaNs, but only correctly set FillValues. I mean I have problems to identify the misuse you mentioned before.
cheers
ralf
| remapCrap.sh (249 Bytes) remapCrap.sh |
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
Sorry Ralph, for the spaghetti of my thought processes. When I said NaN I meant FillValues.
I think I finally have a grasp on what my true problem is...
Operators sel/delgridcell and reducegrid do not have an option to exclude fill values when converting to unstructured.
$ cdo eq zos_Omon_CanESM5_historical_r1i1p2f1_gn_185008.nc zos_Omon_CanESM5_historical_r1i1p2f1_gn_185008.nc mask.nc $ cdo reducegrid,mask.nc zos_Omon_CanESM5_historical_r1i1p2f1_gn_185008.nc test.nc $ ncdump -v,zos test.nc | less
Is there a way to do this?
If reducegrid and/or setgrid,unstructured and/or sel/delgrid had an option to exclude FillValues that would be boss.
RE: Source cell masking in calculating remap weights. - Added by Uwe Schulzweida over 4 years ago
Hi Brendan,
Your demo is not working correctly, because the verbose output of verifygrid is wrong. The cell numbers for the duplicate points are wrong. The coordinates were previously sorted and the output therefore also refers to the sorted array. Since this makes no sense, I have removed this output for the next CDO version.
Cheers,
Uwe
RE: Source cell masking in calculating remap weights. - Added by Ralf Mueller over 4 years ago
Brendan DeTracey wrote:
Sorry Ralph, for the spaghetti of my thought processes. When I said NaN I meant FillValues.
I think I finally have a grasp on what my true problem is...
Operatorssel/delgridcellandreducegriddo not have an option to exclude fill values when converting to unstructured.
They DO have that option. you just have to set the mask values to 0 instead of FillValue. that's why in Uwe's (and my) solution there is
setmisstoc,0as the final step in the mask creation. This sets all missing values to 0. then it works - or at least what I think it is supposed to do.
[...]
Is there a way to do this?
If
reducegridand/orsetgrid,unstructuredand/orsel/delgridhad an option to exclude FillValues that would be boss.
I think setgrid does not take into account any values of the data fields at all. it just sets coordinates
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
Thanks. I did not understand that the mask should not have FillValues. But now:
$ cdo setmisstoc,0 -eq zos_Omon_CanESM5_historical_r1i1p2f1_gn_185008.nc zos_Omon_CanESM5_historical_r1i1p2f1_gn_185008.nc mask.nc $ cdo -v remapcon,global_1 -reducegrid,mask.nc zos_Omon_CanESM5_historical_r1i1p2f1_gn_185008.nc test.nc 2>&1 > log.txt
Gives:
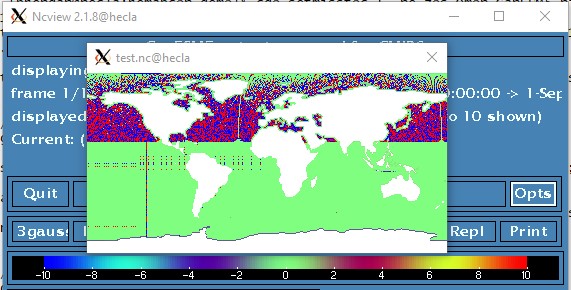
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
Dammit. Please ignore. I forgot to load my environment with the latest version of cdo. Then the last example works perfectly!
RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
Worked beautifully for all samples except IPSL:
$ cdo setmisstoc,0 -eq zos_Omon_IPSL-CM6A-LR_historical_r2i1p1f1_gn_185008.nc zos_Omon_IPSL-CM6A-LR_historical_r2i1p1f1_gn_185008.nc mask.nc cdo(1) eq: Process started cdo(1) eq: Processed 480736 values from 4 variables over 2 timesteps. cdo setmisstoc: Processed 240368 values from 2 variables over 1 timestep [0.06s 41MB]. $ cdo remapcon,global_1 -reducegrid,mask.nc zos_Omon_IPSL-CM6A-LR_historical_r2i1p1f1_gn_185008.nc test.nc cdo(1) reducegrid: Process started cdo remapcon: YAC first order conservative weights from unstructured (120184) to lonlat (360x180) grid cdo remapcon: 2%ERROR: invalid cell Aborting in file clipping.c, line 1295 ...

RE: Source cell masking in calculating remap weights. - Added by Brendan DeTracey over 4 years ago
Which I now see is because the IPSL data has halo. After trimming all is good! Thanks so much for your patience!
RE: Source cell masking in calculating remap weights. - Added by Ralf Mueller over 4 years ago
great - happy to hear that you now got a solution
cheers
ralf
RE: Source cell masking in calculating remap weights. - Added by Feng Wang almost 3 years ago
Hi all,
I had the same problem when trying to regrid some sea ice fraction data from CMIP6 simulations. These files were in Oceanic grid. I wish to remap them to Gaussian grid using the remapcon function. However, it did not work. Following your discussions, I was able to generate a mask from my data. But could not move further. Here is an example of data. Could you give some solutions, please? Thanks.
#I followed this, and it worked.
cdo -L setmisstoc,0 -gtc,-1000 siconc_SImon_MIROC-ES2L_historical_r1i1p1f2_gn_185001-201412.nc mask.nc
#In this step it produced one file which cannot be regridded.
cdo -L reducegrid,mask.nc siconc_SImon_MIROC-ES2L_historical_r1i1p1f2_gn_185001-201412.nc test.nc
Cheers,
Feng
RE: Source cell masking in calculating remap weights. - Added by Estanislao Gavilan almost 3 years ago
Hi Feng,
Did you try to specify the grid.txt file? Like this one
gridtype = lonlat
xsize = 1440
ysize = 721
xfirst = 0
xinc = 0.25
yfirst = -90
yinc = 0.25
you can change the resolution. Then, you just need to typecdo remapcon,grid.txt siconc_SImon_MIROC-ES2L_historical_r1i1p1f2_gn_185001-201412.nc test.nc
RE: Source cell masking in calculating remap weights. - Added by Feng Wang almost 3 years ago
Hi Estanislao,
Thanks a lot. That works well!
Out of my curiosity, why something like below did not work?
cdo remapcon,n32 siconc_SImon_MIROC-ES2L_historical_r1i1p1f2_gn_185001-201412.nc test.nc
Cheers,
Feng
RE: Source cell masking in calculating remap weights. - Added by Estanislao Gavilan almost 3 years ago
Hi Feng,
I am not sure. One of the admins will know for sure. Sometimes the version of the cdo has small bugs, and it does not inform you well about the problem. Uwe said that the 2.0.6 version was going to correct sevEral issues related with remapcon and the attributes of the nc file. Also, it could be that there is something missing in your command or your are working on windows/mac instead of linux. There are many small things that could be the problem. The important thing is you got your grid interpolated! you must celebrate it (hoopefully without baijiu involved)!
Regards,
Estanislao